
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
- Altmetric
GNRA tetraloop-binding receptor interactions are key components in the macromolecular assembly of a variety of functional RNAs. In nature, there is an apparent bias for GAAA/11nt receptor and GYRA/helix interactions, with the former interaction being thermodynamically more stable than the latter. While past in vitro selections allowed isolation of novel GGAA and GUGA receptors, we report herein an in vitro selection that revealed several novel classes of specific GUAA receptors with binding affinities comparable to those from natural GAAA/11nt interactions. These GUAA receptors have structural homology with double-locked bulge RNA modules naturally occurring in ribosomal RNAs. They display mutational robustness that enables exploration of the sequence/phenotypic space associated to GNRA/receptor interactions through epistasis. Their thermodynamic self-assembly fitness landscape is characterized by a rugged neutral network with possible evolutionary trajectories toward natural GNRA/receptor interactions. High throughput sequencing analysis revealed synergetic mutations located away from the tertiary interactions that positively contribute to assembly fitness. Our study suggests that the repertoire of GNRA/receptor interactions is much larger than initially thought from the analysis of natural stable RNA molecules and also provides clues for their evolution towards natural GNRA/receptors.
INTRODUCTION
Stable RNAs including ribozymes, riboswitches and ribosomal rRNAs, fold into their native three-dimensional (3D) architectures through the formation of intricate networks of non-covalent tertiary interactions. Several of these interactions are recurrent long-range self-assembly modules that promote the packing of distant helical regions often separated by several hundred nucleotides (1–9). Among them, A-minor mediated long-range interactions are the most abundant (4,5). They often take advantage of a GNRA tetraloop (R = purine, N = any nucleotide) (1–3,10), in which the two 3' nucleotide positions of the GNRA, typically adenines, can assemble with the minor groove of two G:C base pairs within an RNA helix. This helical receptor is often part of a larger receptor element (1,11,12). Assembly of GNRA-receptor interactions have been shown to contribute to the stability of a molecule's native fold (e.g. (2,13,14), and to affect the speed and accuracy with which the molecule reaches its native state (15). The most abundant GNRA/receptor interactions identified in natural RNAs essentially belong to two main classes: the GYRA/helix and the GAAA/11nt receptor modules (1,2,10,16). These two modules significantly differ in their self-assembly properties, with GYRA/helix interactions being thermodynamically less stable than GAAA/11nt interactions (11,12). This mostly results from the larger number of non-covalent contacts taking place within GAAA/11nt modules. Besides these two classes, few additional natural GNRA- or GNRA-like/receptor modules, including the GUAA/IC3 (17–19), GAAA/Vcr2 (20–22) and L39/H89 modules (23), have been seldomly observed and characterized in a limited number of RNA structural contexts. However, none of these self-assembly modules have been as extensively investigated as the GAAA/11nt self-assembly module (10,11,24–29). In order to further understand the principles governing RNA tertiary interactions and their evolution, various artificial loop-receptors have been isolated by in vitro selection (3,11,30). Using a system based on the mutual recognition of a group I intron and its substrate, Costa and Michel isolated receptors able to recognize GAAA and GUGA tetraloops (3). Interestingly, one of the receptors selected for GAAA displayed preferential binding for GGAA (C7.34) and another receptor selected for GUGA was shown to recognize better GUAA (B7.8), suggesting that many other GNRA/receptor interactions with high affinity and selectivity might exist within the sequence space associated to these types of RNA interactions.
We previously developed a tectoRNA dimer system consisting of two RNA units that synergistically self-assemble through two loop/receptor interactions (31,32). This system allowed investigations of the structural and thermodynamic properties of various GNRA loop-receptor interactions (11,23,33) and also enabled the isolation of two new GGAA receptors by in vitro selection (R1 and R2) (11). Three main structural features were identified as critical for efficient recognition of GNRA tetraloops: the presence of an A-minor interaction submodule, a nucleotide recognition submodule and a platform stabilization submodule (11,23,33). Recently, new GUAA/receptors mimicking the L39/H89 long range interaction from the ribosome were engineered using a purely rational design approach by taking advantage of the modular structural principles of RNA (23). Interestingly, our rationally designed GUAA/S8 interactions were shown to assemble with binding affinities similar to those observed for the GAAA/11nt and GGAA/R1 interactions (11,33) and bore sequence and structural resemblance with the in vitro selected GUAA/B7.8 interaction (23). This strongly supported the view that other GUAA receptors with greater affinity and selectivity than those observed in nature could potentially be isolated by in vitro selection.
With this aim in mind, we modified our previous selection based on the tectoRNA self-assembling system in order to specifically look for potential structural submodules enhancing the stability and specificity of GUAA A-minor mediated interactions. As shown herein, some of the selected GUAA/receptors display structural similarities with known structural modules identified in the ribosome. In addition to isolating novel GUAA/receptor interactions not yet observed in nature, we investigated some of the possible evolutionary pathways and underlying pressures that might have driven the evolution of GNRA/receptor modules toward those commonly observed in natural stable RNAs. While demonstrating that these tertiary modules have a great mutational robustness and display structural plasticity, we also show that they are part of an extended sequence/structural/phenotypic space characterized by a fitness neutral network promoting random drift towards natural long-range interactions.
MATERIALS AND METHODS
Molecular modeling
The bimolecular tectoRNA system used in this study assembles through two GNRA loop/receptor interactions (11,31) according to the atomic model structures available (27,32) (Figure 1 and Supplementary Figure S1). Using the RNA architectonics guidelines (34,35) and SwissPdbviewer (36), we previously built the three-dimensional atomic model for the S8 GUAA/receptor interaction (23) from RNA modules present in X-ray atomic structures (PDB IDs: 1S03, 4V4Q). The three-dimensional (3D) model of the GUAA/R5.58 interaction was derived from the GUAA/S8 model by structural homology modeling using ModeRNA to generate specific, single nucleobase replacements (37). The 3D model of the GUAA/IC3 interaction was built likewise from the GAAA/Vc2 interaction (20). Image rendering and RMSD calculation were performed in PyMOL (38). Proper folding of the secondary structure of each RNA construct was checked using Unafold (39).

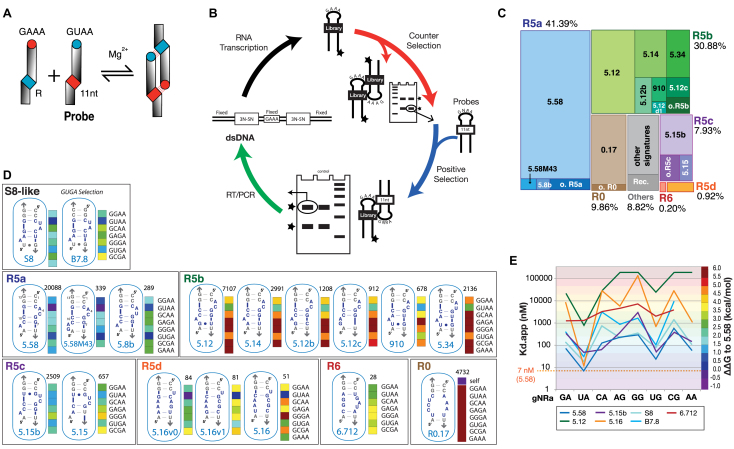
In vitro selection of novel GUAA receptors using a self-assembly tectoRNA system. (A) TectoRNA self-assembly principle (11). R indicates the GUAA receptor region. (B) In vitro selection scheme for the isolation of GUAA/receptors. One typical round of selection is depicted. A total of six rounds of selection were performed (see Materials and Methods). (C) Tree-map visualization of receptor sequences according to their abundance within the G6 HTS pool (total 52168 sequences). Abbreviations: o. stands for other (e.g.: o. R5a, other R5a); Rec., recombination. (D) Secondary structure diagrams corresponding to the most abundant exemplars of each receptor family isolated by SELEX, along with the sequences of S8 and B7.8 receptors, which provide a basis for secondary structure comparison. Selectivity profiles are shown as colored barcodes for receptors tested against GNRA (corresponding ΔΔG are indicated in D). Colors are indicative of the relative stability of each loop/receptor interaction at 10°C in presence of 15 mM Mg(OAc)2 with 0 kcal/mol corresponding to the Kd of GUAA/R5.58 (7 nM). The number above each barcode profile indicates the number of sequence copies identified in the winning G6 tectoRNA pool. (E) Variations of Kds and GNRA selectivity for the most common exemplars of each R5 family of receptors in comparison to S8-like receptors S8 and B7.8. All data were obtained at 15 mM Mg2+ and 10°C as described in Materials and Methods.
TectoRNAs and RNA library synthesis
TectoRNAs (Supplementary Tables S2 and S3) were transcribed with T7 RNA polymerase from PCR generated templates, purified by denaturing polyacrylamide gel electrophoresis (PAGE) and 3′-[32P]pCp labeled as previously described (31,32,40) (see Supplementary Data). RNA molecules were stored at –20°C prior to use. Antisense and primer DNA sequences were ordered from Integrated DNA Technologies (IDT). The initial tectoRNA library comprising of a constant tectoRNA scaffold and a randomized receptor region of 6–9 nts (Supplementary Figure S1A) was obtained by run-off T7 transcription from six DNA library templates generated by PCR, with the random regions being either 3N-3N, 3N-4N, 4N-3N, 4N-4N, 4N-5N or 5N-4N (see Supplementary Data). After separate RNA transcription in presence of alpha-[32P]ATP (Perkin Elmer) and denaturing PAGE purification, the six 32P body-labeled RNA pools were mixed such that the final resulting ‘bank’ pool statistically contained the same ratio of every receptor variants.
In vitro selection
Prior to each selection round, the tectoRNA library was supplemented with the reverse transcription (RT) primer (1:2) in order to prevent its 3′ tail from interfering with the assembly between receptors and probe targets (Supplementary Figure S1A). Each selection round consisted of counter (negative) and positive selections to eliminate pool molecules able to self-assemble in absence of target (rounds 1 and 2) and/or to increase the specificity of recognition for a particular set of GNRA tetraloops versus others (rounds 3–6) (Figure 1B and Supplementary Figure S1B). From rounds 1 to 6, the selection pressure for selective recognition of GUAA versus other GNRA loops was increased in a stepwise fashion by favoring the recognition of GYRA versus GRRA (round 2), GURA versus GRRA (rounds 3 and 4) and GUAA versus GVRA (rounds 4 and 6) (with Y = C or U, R = A or G and V = C,A or G) and by decreasing the concentration of the RNA pools and probes (Supplementary Figure S1B). A typical selection round of selection was as follows (Figure 1B). For the counter selection (red arrow in Figure 1B), the initial RNA library (200 pmol, 15μM for round 1) in presence of the RT primer (as well as probes for rounds 3–6) was submitted to a denaturation step (1 min, 90°C; 3 min, 4°C; 2 min, 30°C) followed by 30 min incubation at 30°C in presence of the association buffer (89 mM Tris-borate pH 8.3, 15 mM Mg(OAc)2, 50 mM KCl final concentration). After a 15 min incubation at 10°C followed by addition of 1/10 volume of gel loading buffer (association buffer with 55% glycerol, 0.05% bromophenol blue and 0.05% xylene cyanol), the tectoRNA library was purified on native 7% (29:1) polyacrylamide gel in association buffer at 10°C in order to eliminate molecules forming RNA complexes. The monomeric RNA species were purified, eluted, ethanol precipitated and then submitted to the positive selection step in presence of the RNA probes of interest (blue arrow in Figure 1B). The RNA pool mixture, including the RT primer and RNA probes, was submitted to the same denaturation/assembly protocol as above. RNA molecules that assembled to the probe target and migrated at the level of a dimer control were purified by native PAGE as described above. Selected RNA molecules were then reverse transcribed using the Improm II RT system (Promega) as specified by the manufacturer and resulting cDNAs were PCR amplified as described in the Supplementary Data (green arrow in Figure 1B). After purification with the QiaQuick PCR Purification Kit (Qiagen), the enriched DNA pool was transcribed (see Supplementary Data) and subjected to an additional round of selection (black arrow Figure 1B). After six rounds of selection, the G6 DNA pool was analyzed by high-throughput Illumina sequencing (Illumina MiSeq, nano output) and the resulting sequence data was processed using EasyDIVER (41) (Supplementary Materials and Methods and Supplementary Table S1).
RNA self-assembly and native gel shift experiments
TectoRNA dissociation constant (Kd) values were determined as previously described (11,23,42,43) (see Supplementary Data for details). Titration experiments were typically performed with equimolar amounts of each tectoRNA at concentrations ranging from 1 nM to 20 μM. After denaturation (2 min, 95°C; 3 min, 4°C; 5 min, 30°C), RNA samples were assembled in presence of association buffer (15 mM Mg(OAc)2 and 89 mM Tris-borate, pH 8.3) at 30°C for 30 min. Samples were then incubated at 10°C for 15 min before native gel analysis [7% (29:1) polyacrylamide gels in association buffer at 10°C] (Supplementary Figure S2). To monitor assembly, one of the monomers contained a fixed amount of 3′ end [32P] pCp-labeled RNA (∼0.25–0.5 nM final concentration). Monomer and heterodimer species were quantified with ImageQuant and dimer formation was plotted against RNA concentration (Supplementary Figure S2). Kd values were determined as the concentration at which half of the RNA molecules were dimerized (see Supplementary Data for details and Supplementary Table S3) and converted to free energy of dimerization ΔG through the formula ΔG = RT ln(Kd), where R is the gas constant and T is the temperature (283 K) (Supplementary Tables S4 and S5). Apparent free energy variation of dimerization for a particular receptor ‘X’ at 10°C (ΔΔG) were derived from the equation, ΔΔG = ΔG(X) – ΔG(reference), with R5.58 WT (Kd = 7 nM) usually taken as the reference. Alternatively, we also used 4000 nM as reference: this value corresponds to the nearest ‘round’ Kd value below which all the tectoRNA helical receptors assemble with their cognate GYRA tetraloops (23).
Chemical probing with dimethylsulfate (DMS)
TectoRNA receptors (2 μM) with an elongated 5′ tail for primer extension were assembled with or without probe (2 μM) as described above in presence of 50 mM HEPES, 100 mM KCl, 15 mM Mg(OAc)2 final concentration. All constructs were tested with the GUAA probe, with the exception of M12 which was tested with the GGAA probe. After incubation, RNA samples were treated with dimethylsulfate (DMS) diluted in 100% EtOH (60 mM final) and reacted for 4 min (monomer) or 8 min (dimer) at room temperature. 2 μl of DMS quench buffer (2.8 M NaOAc, 1 M β-mercaptoethanol) and 200 μl of cold 100% EtOH was added upon completion of the reaction and samples were cooled at –80°C for 15 min and precipitated at 4°C, rinsed twice with 90% EtOH, and dried under vacuum. Samples were resuspended in water and subjected to primer extension using Superscript III RT as described in the Supplementary Information and reference (44). Radio-labeled (32P) reverse transcription products of DMS-modified RNAs were then separated on denaturing 8% PAGE gel and visualized using a Typhon phosphorimager. Sequencing reactions were used as markers and DMS experiments were conducted in duplicate.
RESULTS
In vitro selection of novel GUAA receptors using a self-assembling tectoRNA system
The in vitro selection was modeled after Geary et al. (11), which utilized a self-assembling tectoRNA heterodimer system as shown Figure 1A (see also Supplementary Figure S1). It consists of a tectoRNA library, with a randomized receptor region, able to form heterodimers by assembling to a constant tectoRNA probe (Supplementary Figure S1A). The tectoRNA library was designed so that the randomized region of 9 nts (totalizing ∼6.27 × 105 different sequences) is adjacent to a conserved G:C base pair (bp) doublet located at 11 bp from a GAAA tetraloop (Supplementary Figure S1A). By recognizing the 11nt receptor of the probe, the conserved GAAA loop from the library acts as an anchor to facilitate the selection of novel GUAA binding motifs expected to enhance the formation of A-minor interactions within a structural context similar to known GNRA/receptor interactions. The preservation of the A-minor G:C bp doublet at the level of a minimally randomized receptor region was intended to maximize isolation of A-minor interactions while minimizing isolation of tertiary Watson–Crick base-pairings as previously observed (11). In vitro selection was performed according to the ability of the tectoRNA library to gel-shift on native PAGE upon self-assembly with GNRA probes in presence of 15 mM Mg(OAc)2 at 10°C (see Figure 1B and Materials and Methods). These conditions were chosen as they were originally found to be those allowing Kd measurements of the widest range of GNRA receptors previously studied with the tectoRNA system (11,23,27,31,32,40,42,43). In order to isolate receptors that selectively bind GUAA versus other GNRA tetraloops, the stringency of the selection was increased in a stepwise fashion by subjecting the tectoRNA library to counter and positive selections over a total of six rounds (Supplementary Figure S1B).
The final G6 pool was analyzed by high-throughput sequencing (HTS) (Supplementary Materials and Methods, Supplementary Tables S1AB). After filtering out all sequences unable to fold into the tectoRNA 2D structure, the remaining sequences revealed 1650 different signatures out of which the top 41 and 148 signatures account for 90% and 93% of the total number of sequences (52168), respectively (Supplementary Table S1B). Approximately 91% of the 148 most abundant signatures can be classified into six different families based on their predicted secondary structures: 42 R5a (41.33%), 28 R5b (30.87%), 12 R5c (7.92%), 11 R5d (0.92%), 8 R6 (0.2%) and 8 R0 (9.65%) (Figure 1C). The 27 sequences grouped in the Rec family (1.88%) are the products of recombination events occurring during the RT/PCR step and 12 other sequences are orphans (0.33%) (Figure 1C, Supplementary Table S1B). The remaining ∼7% sequence signatures totalizing ∼1500 different sequences are each represented with a frequency lower than 0.0001. At the exception of R0.17, the top sequence in the R0 family, which self-dimerized with a Kd of 4.2 nM in absence of probes, exemplars from the R5 and R6 families were all shown to assemble with the GUAA probe (Figure 1D and supplementary Tables S1, S3, S4, S5). Interestingly, while none of the isolated receptors had the same sequence as the previously characterized B7.8 and S8 receptors (12,23), the receptor families R5a, R5b and R5c adopt secondary structures resembling those from B7.8 and S8 receptors (Figure 1D). Comparative sequence analysis suggests that the R5a, R5b and R5c families fold as S8-like 2x_bulge modules (12,23) (Supplementary Figure S3). For instance, like the S8 receptor, the R5a family has a CCC:GGG bp triplet at positions X1–3:X11–13 and has the potential to fold as a 2bp_2x_bulge module with positions X6–7 possibly paired to X9–10. The R5b and R5c families can also adopt a 2 bp_2x_bulge module. For example, the sequence signatures of R5.12, R5.14 and R5.12b, which belong to R5b, are closely related to the one of R5.58 from R5a. However, the X3:X11 bp is missing in all R5b exemplars. In the R5c family, this base pair is a G:Y bp instead being a C:G bp (Figure 1D). By contrast, the R5d and R6 families have no apparent structural similarities with S8. The R5d and R6 families possibly fold into an extensive array of Watson–Crick and non-canonical bps, some of them being supported by comparative sequence analysis (Supplementary Figure S3D,E). It is possible that some of the receptors within the pool evolved from some of the most abundant ones. This is likely the case of 5.58M43, which is derived from 5.58 by a nucleotide insertion, and for most single point variants of 5.58 including 5.8b (Figure 1).
Some nucleotide mutations and deletions were acquired within the constant region of the tectoRNA scaffolding during the six rounds of selection (e.g. Supplementary Table S1A), potentially resulting in sub-optimal GUAA binding structures. Therefore, the sequence signatures for each of the selected receptors were encoded within the context of the constant original structural scaffold (Supplementary Tables S2 and S3) to facilitate the characterization and comparison of the binding affinity and GNRA selectivity for all selected receptors (Figure 1D, E, Supplementary Tables S3–S5). The Kd’s for each of the selected receptors were determined and selectivity barcodes and graphs were generated to assess the extent of binding toward GNRA tetraloops in comparison to previously characterized S8-like receptors S8 and B7.8 (23). In some instances, binding for GAAA was also tested when the receptor homo-dimerized. Whereas variations in binding affinity could be observed within each of the R5 families, receptors belonging to the same family of receptors usually shared a similar GNRA selectivity profile (Figure 1D and E). This observation prompted us to hypothesize that receptors with similar structural features might share similar selectivity profiles. Of the selected receptors, R5.58 from the R5a family was the most abundant sequence in the final G6 pool and displayed the greatest affinity for the GUAA loop. In the context of the tectoRNA-dimer system and in presence of 15 mM Mg(OAc)2 at 10°C, the Kd of the GUAA/R5.58 interaction is 7 nM ± 2, which is comparable to the one of the GAAA/11nt receptor interaction, with a Kd of 4.4 nM (11). However, R5.58 is less selective than the 11nt receptor as it displays relatively good binding affinity for all GNRA tetraloops (11). Interestingly, R5a receptors have selectivity profile resembling those from S8-like receptors, with a preference for GURA tetraloops versus all the others. The R5b family favors binding of GUAA versus all the other tetraloops but with a lower affinity than those from other R5 families. It has however a greater selectivity for GUAA versus GUGA than R5a, R5c and S8-like receptors. Interestingly, R5b profiles are similar to the one observed for a variant of S8 lacking the X3–X11 bp (variant S8.24 from reference (23)). Nevertheless, as it will be described later, mutations within the constant scaffold of this particular family of receptors can significantly improve their Kds. R5c receptors have selectivity profiles resembling those of R5a receptors, except that they bind GCAA significantly better than the others. The R5d family binds GUAA (and to a lesser extent GUGA) more selectively than the other R5 families (see Supplementary Tables S3–S5). Whereas R5.58 favors GUAA versus GUGA and GGAA by 3- and 10-fold, respectively, R5.16 favors GUAA versus GUGA and GGAA by 57 and 722 fold, respectively. Finally, R6.712, the most abundant exemplar of the R6 family, which is represented at a very low frequency within the HTS pool, has poor binding affinity for GUAA but still within the range of Kds observed for GYRA/helix receptors. It has also much greater binding promiscuity for any GNRA tetraloops (Figure 1D, E; Supplementary Tables S3–S5).
Mutagenesis of R5.58 indicates that its secondary structure folds as a 2 bp_2x_bulge module
Families R5a, R5b and R5c can fold as GUAA/S8-like receptors, a class of GUAA receptors recently proposed as possible ancestral forms of the L39/H89 ribosomal universal interaction (23). All R5a, R5b and R5c sequences can form a secondary structure similar to the 2 bp_2x_bulges of S8 and B7.8 receptors (Figure 1D). However, they can also potentially adopt alternative conformations. As shown in Figure 2A, no less than four different 2D structure models can be proposed for R5.58, the top selected receptor which accounts for more than one third of all selected sequences. In order to find out whether R5.58 and S8 had a common 2D structure, we performed an extensive, systematic point mutation analysis of R5.58 and tested the resulting variants for their ability to recognize GNRA tetraloops at 15 mM Mg2+ and 10°C (Figure 2B and C, Supplementary Figures S4, S5, Supplementary Tables S3–S5). As shown in Figure 2B and Supplementary Figure S4, single and double mutants at positions X3:X11, X6:X10, X7:X9 and X7.1:X7.2 strongly support the existence of Watson–Crick base pairs at these positions. While single mutants at positions X3:X11, X6:X10 and X7:X9 are usually more disruptive than the wild type, compensatory double mutants maintaining Watson–Crick bps significantly rescue binding affinity for GUAA (Supplementary Figure S4). This corroborates that R5.58 likely adopts the same secondary structure as S8 when bound to GUAA. Positions X1:X13, X2:X12 and X3:X11 bps are involved in the formation of the A-minor interaction with the GNRA tetraloop target (11,23). As expected, X2:X12 and X3:X11 bp, which are directly involved in the formation of the A-minor interaction with the two last nucleotides of the GNRA (11,23), display a stronger preference for the wild type sequence than the X1:X13 bp, which is not directly in contact with the GNRA tetraloop. The C1:G13 bp can easily be substituted by any Watson–Crick bps without much loss of affinity for GUAA. Likewise, the closing U7.1:A7.2 bp can be easily substituted by a G:C or G:U bp without significant loss of binding affinity for GUAA.

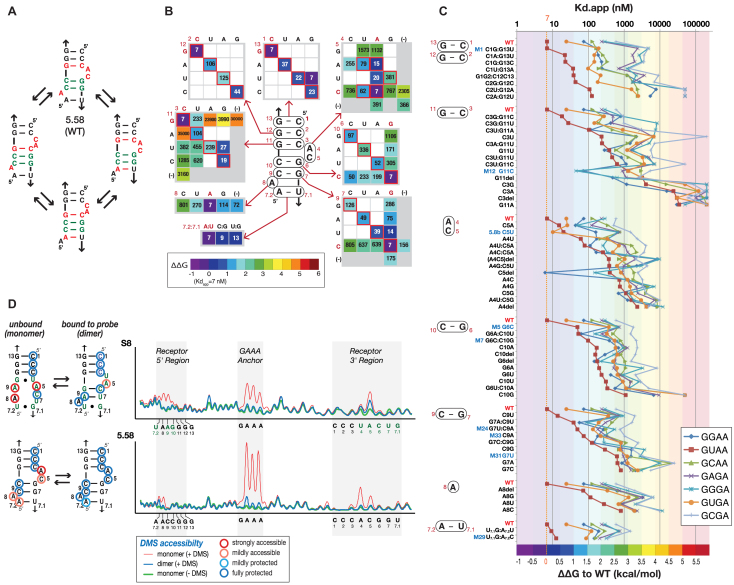
Characterization of the structure of R5.58 by mutagenesis, gel-shift assays and DMS probing. (A) Possible alternative secondary structures adopted by R5.58. The working 2D model is shown on the top. Nucleotide positions in red are those involved in alternate structures. (B) Equilibrium constants of dissociation Kds (in nM) obtained by self-assembly of R5.58 tectoRNA variants with the GUAA probe corroborate the 2D structure of R5.58 as depicted. Values boxed in red correspond to the most common expected bps (cis W:W bp for X1:X13, X2:X12, X3:X11, X6:X10 and X7:X9 variants; cis HG:SG bps for X4:X5 variants). (46). Kd values for the GUAA probe are in nM and the color code corresponds to the ΔΔG values shown in the box. (C) Binding affinity and GNRA tetraloop selectivity for all R5.58 tectoRNA variants tested. Some mutations at positions X3:X11 and X4:X5 can lead to change of GNRA selectivity, suggesting a direct involvement of these positions in the recognition of the second and third position of the GNRA. (D) DMS modification profiles and corresponding secondary structures for the S8 and R5.58 receptors in the bound or unbound states within the tectoRNA context (see also Supplementary Figure S9). Receptors in the bound state were probed in presence of the GUAA probe. Color codes used for DMS accessibility: see the box at the bottom right. All data were obtained at 15 mM Mg2+ and 10°C as described in Materials and Methods.
Comparative homology modeling of the R5.58/GUAA interaction
The RNA binding site of ribosomal protein S8 within bacterial 16S rRNAs folds as a ‘double-locked’ bulge (2 bp_2x_bulge) module, which involves two nucleotide (nt) platforms (type 1a and type 2a) (see Figure 3) (23). When bound to the S8 protein, the type 1a nt platform serves as a stacking platform for recognizing amino acids 105–106 (Thr105–Ser106 in E. coli) of the S8 protein (45) (Figure 3A). The S8 2x_bulge module was previously demonstrated to be a remarkable receptor for GNRA tetraloops, most especially for GUAA (23). Herein, we used the detailed atomic model of the S8/GUAA interaction for homology modeling of the R5.58/GUAA interaction (Figure 3BC and Supplementary Figure S6). The intricate network of H-bonds and stacking contacts of the S8 receptor can be generated likewise with the R5.58 sequence despite the fact that the R5.58 sequence differs at eight nucleotide positions (out of a total of 15) from the S8 sequence (Figure 3). The root-mean-square deviation (RMSD) between the ribose/phosphate backbone of the two models is <1 Å, emphasizing their remarkable isostericity (Figure 3D). While the two last adenines of the GUAA tetraloop form an A-minor interaction with the second and third base pair of the receptor (11), the second nucleotide of the tetraloop is involved in a base-base stacking with the proximal type 1a nt platform and could be in partial or indirect contact with the third base pair (C3:G11) of the receptor (Figure 3C and Supplementary Figure S6CD). Both type 1a and type 2a platforms involve an AC cis Hoogsteen:Shallow groove (HG:SG) bp (Supplementary Figure S6EF), which is one of the most abundant cis HG:SG bp identified in ribosome atomic structures (46). Typical of S8-like 2x_bulges, nucleotides A4 and A8 stack on one another and lock the proximal type 1a and distal type 2a platforms to one another through two WC-2′OH contacts (Figure 3C and Supplementary Figure S6G).

![Structural homology between the GUAA/R5.58 and GUAA/S8-like interactions. 2D diagrams and 3D atomic structures for (A) the RNA binding site of the S8 ribosomal protein from the E. coli 16S rRNA [PDB_ID: 4V4Q] (45,49), (B) the GUAA/S8 interaction (23) and (C) the GUAA/R5.58 interaction. Numbering and non-canonical tertiary contacts are indicated. The nomenclature for base pairs and tertiary contacts is according to references (6,12,71). The A-minor is in violet and type1a and type 2a nt platforms are in yellow and orange, respectively. The interacting stacked loop nucleotide and amino acids are in red. (D) Structural superposition of the three 2x_bulge modules.](/dataresources/secured/content-1765949017427-0cbba233-96bd-4d3b-ace5-1e10fced7083/assets/gkab021fig3.jpg)
Structural homology between the GUAA/R5.58 and GUAA/S8-like interactions. 2D diagrams and 3D atomic structures for (A) the RNA binding site of the S8 ribosomal protein from the E. coli 16S rRNA [PDB_ID: 4V4Q] (45,49), (B) the GUAA/S8 interaction (23) and (C) the GUAA/R5.58 interaction. Numbering and non-canonical tertiary contacts are indicated. The nomenclature for base pairs and tertiary contacts is according to references (6,12,71). The A-minor is in violet and type1a and type 2a nt platforms are in yellow and orange, respectively. The interacting stacked loop nucleotide and amino acids are in red. (D) Structural superposition of the three 2x_bulge modules.
The modularity of R5.58 is compatible with the one of S8-like receptors
The structural modularity of the R5.58 and its compatibility with the S8 GUAA receptor were further investigated by looking at the way mutations affect GNRA binding affinity and selectivity (Figure 2C, Supplementary Figure S5).
Analysis of the A-minor submodule
Mutations were introduced into the X1–2:X12–13 bp doublet known to participate in the formation of the type I/II A-minor interaction with the last conserved adenine position of the GNRA. While altering the affinity for tetraloops, most of these mutations do not drastically alter the relative selectivity of the receptor to the seven GNRA tetraloops (Figure 2C). For instance, most variants bind preferentially GUAA followed by GUGA. However, C1G:G13U (M1) significantly increases GUAA specificity while retaining the same Kd as R5.58 (See also Supplementary Figure S2). Several mutations in the X3:X11 bp, which is involved in a type II A-minor interaction with the purine (R) in the third position of the GNRA, altered not only the affinity but also GNRA selectivity. Several variants (e.g. C3U:G11A, G11U, G11C, G11del, C3G) displayed better affinity for GGAA and/or GGGA than GUAA. Most noteworthy is G11C (M12) which favors GGGA (45 nM) and GGAA (72 nM) versus GUAA (1285 nM) by 18- and 28-fold, respectively. Other variants, while still recognizing best GUAA, did not bind GUGA as well as other GNRA (e.g. C3U, C3A:G11U) (Figure 2C). This suggests that the X3:X11 bp is in close proximity to the second position of the tetraloop (Supplementary Figure S6D). Interestingly, this bp within the S8 receptor context was also found to contribute to GUAA selectivity (23).
Analysis of the type 1a platform X4:X5 from the proximal submodule
In the crystallographic structure of receptor S8 (45), nucleotides at positions X5 and X4 are involved in a cis HG:SG bp, creating a type 1a nucleotide platform unto which the second position of the GUAA could potentially stack (23) (Figure 3 and Supplementary Figure S6EF). The existence of a similar bp in the context of R5.58 is supported by mutagenesis analysis (Figure 2B, Supplementary Figures S5B and S7). For instance, several of the X4:X5 variants which bind best GUAA are also those with nucleotide combinations corresponding to the most abundant cis HG:SG bps identified in atomic structures (46) (Supplementary Figure S5B). As such, four out of the five most abundant combinations of cis HG:SG bps (AC, AA, UC and UA platforms) are shown to recognize best the GUAA tetraloop with Kds below 100 nM. By contrast, other combinations (CC, GC, AG and UG), which are not usually observed in naturally occurring RNAs, resulted in significant loss of binding affinity for GUAA (Figure 2B, Supplementary Figure S7) and even contributed negatively to binding when compared to the X4:X5 deletion variant R5.58M41, which behaves as a GYRA/helix receptor interaction (Supplementary Figure S7). Several of the GNRA selectivity profiles of X4:X5 variants in the R5.58 context are similar to those in the S8 context (see AC, AA, CA, GU and double deletion mutants) but some differences can also be observed for combinations AU, UC, UA, AG and UG (Supplementary Figure S7). The AU platform leads to enhanced binding for GUGA with respect of GUAA in the R5a structural context (Selected receptor R5.8b) but it significantly destabilizes GURA binding in the S8 context (Supplementary Figure S7). In the context of S8, the UC and UA platforms confer greater selectivity of recognition towards GUAA versus GGRA tetraloops than they do in the R5a context. In the context of R5a, AG and UG variants recognize better GGAA than GUAA whereas the opposite trend is observed in the context of S8 (Supplementary Figure S7). This data suggests that sequence variations within the nucleotide platform X4:X5 modulate binding affinity and GNRA selectivity. The C5del variant, which favors GGAA (6.2 nM) versus GUAA (391 nM) by 63-fold is another noteworthy example emphasizing the importance of the proximal bulge for GNRA selectivity. Like for the X3:X11 bp, mutations at positions X4:X5 support a direct involvement of the X4:X5 platform of R5.58 in the binding/recognition of the third position of the GNRA tetraloop. As previously observed for the S8 receptors from E. coli and T. Thermophilus 16S rRNAs (23), the nature of the sequence of the structural context surrounding the X4:X5 platform can have differential effects on GNRA selectivity and affinity, suggesting that some X4:X5 variants might fold into alternative structures when in presence of a particular GNRA tetraloop. The X6:X10 bp unto which X5 is stacked, is validated by Watson–Crick compensatory changes but can tolerate mismatches (e.g. variant M5 (G6C) in Figure 2B, Supplementary Figures S4, S5B) without affecting GUAA selectivity.
Analysis of the distal type 2a submodule
The mutation analysis within positions X7:X9, X8 and X7.1:X7.2 indicates that these positions are not as critical for GNRA recognition and selectivity as those involved in X3:X11 and X4:X5 (Figures 2C and S5C). As suggested by the R5.58 and S8 structural models, none of these positions makes a direct contact with the GNRA (Figure 3). Interestingly, in the R5.58 context, deleting A8 is not as detrimental for GUAA affinity as the A8U and A8C substitutions, and to a lesser extent, the A8G substitution (Figure 2, Supplementary Figure S5). This supports the involvement of A8 in a tertiary contact. According to the models, position X8 forms a cis HG:SG bp with position X9 of the X7:X9 bp (Figure 3). As such, an adenine in X8 is best suited for forming this contact with a G:C bp in X7:X9. Moreover, slightly improved binding affinity for GUAA is observed when the A8G substitution is associated with a U:A bp for X7:X9 (5.58M30, Supplementary Figure S5C), which is in good agreement with the formation of a G:U cis HG:SG bp. Additionally, combinations of nucleotides that potentially favor the formation of an A8A9 HG:SG bp within the context of the S8 and R5.58 receptors are among those that contribute best to GUAA binding (e.g S8, R5.58M28, R5.58M33, R5.58M24 Supplementary Figure S5C).
In summary, the mutation analysis data supports the hypothesis that R5.58 folds as a ‘S8-like’ 2x_bulge module when bound to GUAA. Overall, our results also highlight the remarkable robustness of this receptor upon mutations. For instance, 41%, 85% and 95% of all the R5.58 variants have Kds for GUAA below 100, 1000 and 4000 nM, respectively (Supplemental Table S3, Supplementary Figure S8). The high sequence plasticity of the R5a family of receptors suggests that these receptors are highly evolvable and might allow traversal of the local sequence/structural space with potential structural changes.
DMS chemical probing corroborates that R5.58 folds as a S8-like receptor
The structures of the S8 and R5.58 receptors were investigated further by dimethyl sulfate (DMS) chemical probing within the tectoRNA context in the presence or absence of GUAA target (Figure 2D and Supplementary Figure S9). DMS reacts with Watson–Crick positions N1 of adenines and N3 of cytosines when accessible to the solvent (44). It is therefore particularly well suited for probing the structure of R5.58, which is comprised of 9 As and Cs over a total of 15 nts. As seen in Figure 2D, tectoRNAs bearing the S8 or R5.58 receptors have their GAAA tetraloop anchor fully accessible to DMS modification in the unbound state (without GUAA probe) and fully protected in the bound state, attesting that R5.58 and S8 tectoRNAs are fully assembled to the GUAA probe in the bound state. For S8, the formation of the central Watson–Crick bp doublet (X6–7:X9–10) is only formed by induced fit upon binding of GUAA. Additionally, the bulging nucleotides involved in the formation of the type 1a and 2a nt platforms become protected towards DMS only in the bound state. For R5.58, the nucleotides involved in the formation of the secondary structure of the 2x_bulge module are protected in both unbound and bound states, suggesting that its secondary structure is more stable than the one of S8. However, like for S8, the bulging nucleotides of R5.58 (A4C5 and A8) are only protected towards DMS in the bound state, suggesting an induced fit mechanism for the formation of the proximal and distal nt platforms upon binding of GUAA (Figure 2D) (23). This protection pattern is likely due to the cross-strand hydrogen bonding occurring between the 2′-hydroxyls of A4 and A8 and their respective N1 positions, one of the characteristic structural signatures that make up the ‘S8-like’ fold (Figure 3 and Supplementary Figure S6G). Moreover, N3 protection of C5 likely stems from the stacking interaction with the U residue in the GUAA tetraloop, indicating the influence of the proximal type 1a platform on tetraloop binding.
Structural and phenotypic plasticity of the R5a family of receptors
Additional R5.58 variants were subjected to DMS probing to investigate the effect of single point mutations disrupting key WC bps (M12 G11C, M5 G6C, M31 G7U in Supplementary Figure S9). G11C, which disrupts the X3:X11 bp involved in the recognition of the GUAA tetraloop, changes the receptor selectivity from GURA to GGRA and was therefore studied in presence of the GGAA rather than GUAA probe (see Supplementary Figure S9). Nucleotides involved in the formation of the M12 receptor are mostly accessible to DMS modification in absence of GGAA but they become protected in the bound state: the overall modification pattern for M12 is compatible with the induced fit formation of a S8-like receptor. In the case of M5 G6C and M31 G7U, receptor nucleotides remain accessible to DMS in both bound and unbound states, suggesting that these receptors are structurally metastable and/or that the equilibrium of dissociation between the receptor and tetraloop is more dynamical (Supplementary Figure S9). This is in part corroborated by the fact that the GAAA/11nt anchor interactions of these tectoRNAs are not fully protected from DMS when in the bound state. DMS modification patterns suggest that the M31 receptor adopts an alternative fold from the canonical S8-like secondary structure (see Supplementary Figure S9). Instead forming bps X3:X11, X6:X10 and X7:X9 like in R5.58 and S8, the mutation G7U induces the formation of bps X5:X11, X6:X9 and X7:X8, with X10 and X3 × 4 bulging out: this can also explain the decrease of GUAA binding affinity of M31 (Kd 637 nM) versus R5.58 (7 nM). We also investigated the DMS modification pattern of the variant M28, which was initially designed as a hybrid between S8 and R5.58 as it comprises the type 1a platform submodule from R5.58 combined to the type 2a submodule of S8 (Supplementary Figure S9). M28 is four mutations away from both R5.58 and S8. Whereas M28 has a binding affinity for GUAA of 2.6 nM and has a GNRA selectivity profile resembling the one from R5.58, its DMS modification patterns are remarkably different from those of R5.58 and S8. Upon binding of GUAA, modification patterns of M28 suggest the induce fit formation of an alternative 2x_bulge module that might involve formation of bps X5:X11, X6:X10 and X7:X8, with nucleotides A9 and C3A4 bulging out (Supplementary Figure S9). As such, the 2D structure of M28 is closer to the one of the R5b family of receptors and only three mutations away from R5.34. A detailed three-dimensional model of the GUAA/M28 interaction is not available yet.
In summary, while our data supports structural homology between R5.58 and S8, the fact that the M28 structure is significantly different from the one of R5.58 and S8 not only highlights the remarkable sequence plasticity of this family of receptors but also its structural plasticity.
Exploring the sequence/structure/phenotype landscape associated to R5 receptors
Considering the remarkable plasticity of receptor R5.58 (or 5.58), we hypothesized that the mutational robustness of R5.58 is particularly suited to explore the sequence/structure/phenotype space associated to GNRA/receptor self-assembly and that possible mutational pathways might link the R5a family to the other families of selected receptors. In order to test this hypothesis, we generated a sequence space network connecting S8 and the most abundant R5 receptors (total of 14) through continuous point mutational pathways (Figure 4 and Supplementary Figure S10). Shortest mutational pathways were identified by determining the Levenshtein distances separating selected and S8 receptors from one another (Supplementary Figure S10AB): the Levenshtein distance (L) separating two receptor sequences defines the shortest number of changes (including substitutions, insertions and deletions) to transform one sequence into another. While the most abundant selected receptors were readily connected by one-point mutation (e.g. 5.58, 5.58M43 and 5.8b; 5.12, 5.12b, 5.12c and 5.14; 5.34 and 910), intermediate variants were picked within the G6 pool or rationally designed in order to connect the most closely related R5 sequences and to minimize the number of intermediates necessary to link all receptors. Previously characterized S8 and R5.58 variants were also taken into consideration in the build-up of the sequence space network in order to minimize the number of new variants to be tested. The final network comprises 73 nodes (31 R5 and S8 receptors and 42 intermediates) linked by 107 edges of Levenshtein distance equal to 1. All the receptors were tested for their affinity and selectivity towards GNRA tetraloops (except GAAA) in order to create the sequence/phenotype fitness landscape displayed in Figure 4 (see also supplementary Figure S10C). The ‘fitness’ criterion was calculated as the variation of binding free energies ΔΔG for GNRA/receptor interactions using as a reference the Kd value of 4000 nM, which corresponds to the nearest ‘round’ Kd value below which all the tectoRNA helical receptors assemble with their cognate GYRA tetraloops (23). Therefore, receptors deemed more fit were those enhancing recognition of GNRA tetraloops with respect of regular helical receptors.

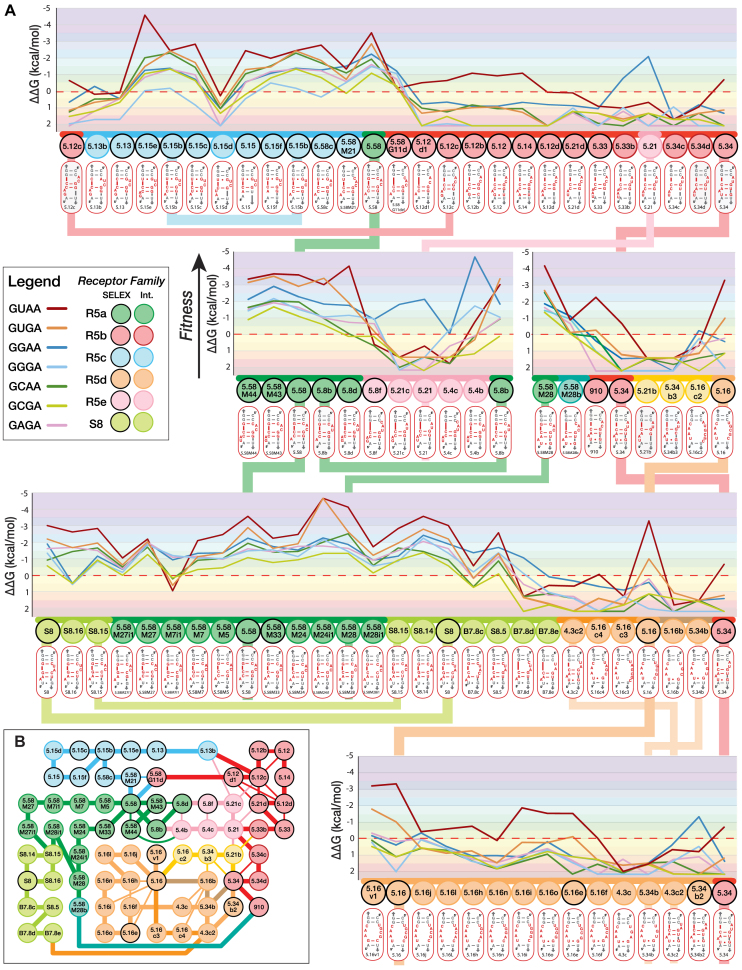
Fitness landscape of a sequence mutational network connecting R5 and S8 receptors. (A) Loop/receptor binding affinity (ΔΔG) within the tectoRNA context is taken as the fitness phenotype measured in comparison to the binding affinity of GNRA tetraloops to regular helical receptors: 0 kcal/mol corresponds to a Kd value of 4000 nM that was chosen as the reference threshold because it corresponds to the nearest round Kd value below which all the tectoRNA helical receptors assemble with their cognate GYRA tetraloops (23). All ΔΔG values are calculated with respect to the 4000 nM Kd activity threshold, at 10°C (See Supplementary Table S5). The fitness y-axis is in a reversed orientation in comparison to Figure 1E. Receptor families corresponding to selected receptors (SELEX) and intermediate variants (int.) are color-coded as indicated in the box legend. The secondary structure of each receptor is also indicated. (B) Sequence mutational network for all receptors shown in (A). Selected R5 receptors are linked through some of the shortest Levenshtein distances (L) (see Supplementary Figure S10A). Connecting edges (L = 1) are color-coded according to receptor families. Thick edges correspond to mutational pathways shown in the fitness landscape in (A). Thin edges (L = 1) connect receptors (nodes) that are not shown next to one another in the fitness landscape.
The present sequence/phenotype landscape accounts for a small portion of the full sequence space and mutational pathways associated to these receptors. Nevertheless, it highlights interesting features. The fitness landscape is rugged but, at the exception of 5.16, all the selected receptors can be connected through mutational pathways with ΔΔG below 0.0 kcal/mol when compared to GYRA helical receptors (Figure 4; or ΔΔG ranging between –0.5 and 4 kcal/mol in Supplementary Figure S10C): out of the 73 receptors, only one out of six (one out of three for intermediates) did not bind efficiently any of the GNRA tetraloops. Family R5a which includes R5.58, is connected through its sequence/phenotype space to families R5b, R5c and S8. This suggests that random mutational walk within the fitness landscape associated to these families of receptors is possible through evolution. Most of these receptors can retain a secondary structure compatible with the formation of a 2x_bulge module. Preferred selectivity for GURA versus the other GNRA can be maintained for most sequence variants within these families at the exception of few mutational transitions which led to preferential binding to GGRA. For instance, it is possible to walk through continuous point mutations from R5.58 to S8 and from R5.58 to 5.15 without significant loss of affinity and GNRA specificity (Figure 4). Interestingly, mutational transition involving in/del mutations are more prone to change of specificity than substitution mutations (Figure 4 and Supplementary Figure S11). For instance, the deletion of G11 (G11del) in different receptors usually led to the same change of specificity from GURA to GGRA, emphasizing the importance of the X3:X11 bp and the X4:X5 platform for the specific recognition of the GNRA tetraloop (see Supplementary Figure S11A). Nevertheless, the sequence context of the distal platform can also affect differently a particular mutation outcome. This is illustrated by the subsequent deletion of C3 (C3del) which can either retain or reverse back GNRA selectivity (Supplementary Figure S11A). The addition of G8.1 in the distal bulge of 5.8b and 5.15b, which leads to 5.8d and 5.15e, respectively, increases the selectivity and affinity for GUAA (Supplementary Figure S11B) but no significant changes are observed for 5.58M43 versus 5.58. However, additional mutations in the distal platform region can have unexpected effects on both affinity and selectivity. For instance, the G8.1U point mutation in the distal platform of receptor 5.8d (Kd = 2.7 nM) leads to variant 5.4b, which is the best GGRA receptor (Kd = 1nM) characterized in this study; the same mutation within 5.58M43, leading to 5.58M44, does not change selectivity. Similarly, base substitutions in the closing distal bp X7.1:X7.2 had different effects depending on the sequence contexts (Supplementary Figure S11C). Receptors 5.58M24 and 5.58M7 have similar GNRA selectivity profiles. However, the substitution U7.1G in the 5.58M24 context dramatically improved binding affinity for GURA (5.58M24i1) whereas it led to a change of selectivity toward GGRA in the 5.58M7 context (Supplementary Figure S11C). Likewise, the mutational transition G10C + C6G (or G6C + C10G), which corresponds to the transversion of C6:G10 into G6:C10 (or vice versa), leads to a transitory change of loop selectivity in the context of S8 but not in the context of R5.58 (Supplementary Figure S11D). These are clear indications of epistasis, where the effect of one particular mutation is dependent on the sequence context of a particular receptor. Whereas families R5a, R5c and S8 preferentially bind GUAA, most variants still display significant binding affinity for other GNRA tetraloops. Therefore, even in the case of change of loop selectivity, mutations occurring in these GNRA/receptor interactions enable continuous random walk by maintaining better Kds than most GNRA/helical receptors.
Several mutational pathways with the shortest Levenshtein distances separating 5.34 and 5.16 from the other selected receptors were tested but none of them allowed 5.34 and 5.16 to be connected through intermediates with Kds below 4000 nM for any GNRA. Nevertheless, receptor 5.34, which belongs to the R5b family, could be connected (via 5.58M28) to the R5a family through a mutational pathway of longer Levenhstein distance. Likewise, some pathways linking 5.16 to the other R5 receptors with Kds below 4000 nM are likely to exist as only few of these possible pathways have been tested in the present study. As a matter of fact, 5.16 and its variants are connected to newly selected GGAA receptors that are part of an even larger sequence/phenotype landscape (Zakrevsky, Calkins and Jaeger, in preparation).
Synergetic effect of scaffold mutations for the self-assembly of R5b receptors
The relative abundance of the selected receptors in the HTS pool is likely representative of their fitness, the receptors with highest GUAA affinities being more abundant than the others. However, despite the fact that all R5b variants are able to be connected to the R5a family, R5b receptors had lower binding affinities for GUAA than initially expected. Yet, the length of the A-minor stem is important for optimal assembly of the tectoRNA dimer system (23,31,32) and because of their shortened A-minor stem, the R5b receptors assemble less well than the other R5 receptors within the tectoRNA context (Figure 4). Interestingly, analysis of the R5b sequences indicates that most of them have at least one extra nucleotide inserted within the stem separating the two interacting GNRA/receptor modules (Supplementary Table S6 and Figure S12A). This suggests that these scaffold mutations, introduced within the constant region during the SELEX process, might have positive synergetic effects on the tectoRNA assembly of the R5b family of receptors. As expected, scaffold variants of 5.12, 5.12c, 5.12d, 910, 5.21, 5.34 and 5.33 have significantly lower Kds (and better fitness), which equate to those from the R5a family (Supplementary Tables S3,S4,S5 and Supplementary Figure S12). This binding improvement can be >100 fold for GUAA (Supplementary Figure S12BC). R5b scaffold variants also display greater GUAA selectivity than 5.58. These data stress the positive contribution of nucleotide positions located far away from the self-assembling interactions and also explain why the R5b family is the second most abundant family of selected receptors. Consequently, the pathways linking 5.34 to the R5a family and 5.33 to the other R5b receptors are significantly smoothed by the positive synergetic contribution of these scaffold mutations within the tectoRNA context; all R5b variants have Kds well below 4000 nM for all GNRA tested (Supplemental Figures S10D and S12D).
Synergetic scaffold mutations could likewise improve many of the suboptimal receptor intermediates and selected variants of the R5b family and R5e family, which includes 5.21 (Supplementary Figure S12). In summary, the resulting fitness landscape associated to these receptor families correspond to an extended rugged neutral network connecting most of the selected receptors through neutral drift (or nearly neutral mutational steps). In the tectoRNA system, synergetic scaffold mutations significantly smooth the ruggedness of this neutral network by enhancing the binding fitness of several selected receptors and variants in comparison to regular GNRA/helix receptors.
Recreating possible evolutionary paths between the GUAA/5.58 and natural GNRA/receptor interactions
The data presented above suggests that the sequence/phenotype space associated to the R5 receptors families might also be linked to naturally observed receptors, including the GAAA/11nt, GYRA/helix, GUAA/IC3 and GAAA/Vc2 interactions. The GAAA/11nt and GYRA/helix receptors are the two most widespread classes of GNRA receptors identified in natural RNA molecules: both have been observed in numerous X-ray structures (Figure 5; See Supplementary Table S1 from Ref. (11)). While the 11nt recognizes with high affinity and selectivity the GAAA tetraloop (Kd = 4.4 nM) (11), the GYAA/helix receptors, with Kds ranging between ∼760 and ∼4300 nM for GUAA, are not as thermodynamically stable and display a preference for GUAA, GCAA and GUGA, with GGAA, GGGA and GAAA being recognized the least (Supplementary Tables S3–S5). As previously reported (1,11,12), when the type I/II A-minor interaction involves a CC:GG bp doublet, it typically prefers GYAA versus GYGA while the opposite is true when the helix doublet is CU: AG. The difference in selectivity is however moderate, with Kds varying by <3–4-fold (Supplementary Table S3). As shown herein, the CU:GG and CC:AG bp doublets do not bind to any GNRA in the experimental conditions tested, suggesting that a transition mutation from GYAA/helix to the GYGA/helix receptor is somewhat energetically disfavored (see hGYGAb in Figure 5C and hGYGAc in Supplementary Table S3). Overall, GYAA/helix interactions are mutationally robust as the bp positions below the A-minor doublet can tolerate mismatches (e.g hGYAAj, hGYAAi, hGYAAg) as well as nucleotide insertions (e.g. hint3, hint4, R5.58M41, IC3g, IC3c) (see Figure 5C and Supplementary Table S3). The IC3 (11,17,19) and Vc2 (22) receptors, which are more sparsely found in stable RNAs, belong to the same class of receptors as they have similar sequence signatures with similar secondary structures. As the X-ray structure of the GAAA/Vc2 interaction is known (20), a 3D model of the GUAA/IC3 interaction was obtained by structural homology modeling from the Vc2 atomic structure (Figure 5B). Kd values for the GUAA/IC3 and GAAA/Vc2 interactions are 30 nM and 134 nM, respectively (Supplementary Table S3). Both receptors have been proposed as evolutionarily linked to the 11nt receptor (17,22) and this is strongly supported by our results displayed in Figure 5. In fact, the sequence/phenotype landscape associated to natural classes of receptors shows that it is possible to move through the sequence space from one natural receptor to another with binding energy remaining below ΔΔG = 0 kcal/mol. Interestingly, the R5.58 receptor (and related receptors) can be readily connected to the more distant 11nt, IC3 and Vc2 receptors via GYRA/helix natural receptors without entailing dramatical loss of binding energy (Figure 5). Deletion of the bulging nucleotide involved in the formation of nt platforms in the R5.58 receptor leads to GYAA/helix receptors, which preferentially bind GUAA, albeit with reduced affinity and selectivity in comparison to the R5a family. The continuous mutational pathway existing between the GUAA/helix interaction (hGYAAh) and the GAAA/11nt interaction necessitates change of loop specificity from GUAA to GAAA. This is possible because some receptor intermediates have more promiscuous binding of GNRA tetraloops and share GNRA selectivity patterns similar to those from the IC3 and Vc2 natural receptors. In summary, the R5a, R5b and R5c families of receptors are connected to the natural receptors through a neutral fitness landscape that highlights their mutational robustness and structural plasticity. In nature, these characteristics can drive their evolution toward the more selective GAAA/11nt receptor.

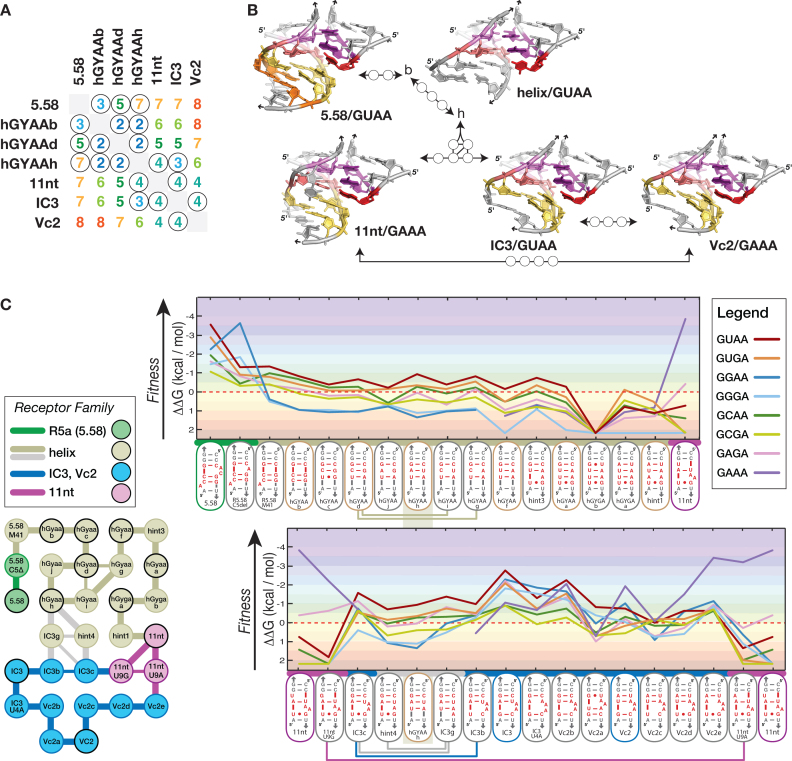
The GUAA/R5.58 receptor is linked to the sequence/structure/phenotype landscape of natural GNRA/receptors. (A) Levenshtein distances connecting R5.58 to natural receptors. The circled values in the table correspond to the shortest distances tested. (B) Comparison of the 3D structures of the GUAA/R5.58 receptor and the natural receptors GAAA/11nt, GYRA/helix, GUAA/IC3 and GAAA/Vc2. (C) (left) Sequence mutational network connecting R5.58 to the natural receptors and (right) corresponding fitness landscape. Connecting edges (L = 1) and receptor nodes are color-coded according to receptor families (same legend as Figure 4B). All ΔΔG values are calculated with respect to the 4000 nM Kd activity threshold, at 10°C (see legend of Figure 4A and Supplementary Table S5).
DISCUSSION
Knowledge of the accessible sequence space or designability associated to natural RNA modules can be seen as a 3D structural code that can help understand possible evolutionary constraints as well as improve the prediction and the rational design of novel RNA architectures. Herein, we have used a self-assembling tectoRNA system for the direct selection of novel GNRA-binding receptors promoting helical packing. In contrast to previous in vitro selection systems aiming at isolating receptors for GAAA, GUGA and GGAA tetraloops (3,11), the selection was designed for isolating structural features possibly enhancing the formation of A-minor interacting modules between a GUAA tetraloop and a helical stem. Several new families of GUAA-binding receptors were isolated and were characterized to have binding affinities comparable to other selected and natural GNRA/receptors. Our initial library, which was randomized at only nine positions (6.27 × 105 different variants), represented only 1.34% of the sequence space necessary for encoding all the possible types of RNA internal loops identified in the ribosome. Therefore, our search was far from being exhaustive and missed known structural modules contributing to the stabilization of GUAA mediated A-minor interactions. For instance, our initial library design did not include asymmetrical internal loops in which the nucleotide length of one side exceeded the other by two or more nucleotides, which prevented isolation of IC3-like receptors. Moreover, it did not comprise modules with sizes above 13 nts, which impeded the isolation of the S8 receptor and possibly other receptors requiring mutations in the closing base pairs X7.1–X7.2 (Supplemental Table S1B). Nevertheless, due to incorporation of mutations and recombination events during the in vitro selection process, several receptor intermediate variants, which were not initially present in the G0 library, could be isolated in the G6 pool, albeit at a very low level (Supplemental Table S1B).
Remarkably, three out of the five selected families of GUAA receptors present structural homology with the S8-like ‘double locked bulge’ module previously identified in the ribosome and for which atomic crystallographic structures are available (12,23,45,47) (Figure 3, Supplementary Figures S3 and S6). Double locked bulges (2x_bulge) constitute a widespread class of structural modules that are often in interactions with RNA loops and/or proteins in stable natural RNA molecules (12,23,47,48). In a previous study (23), the S8 module was rationally designed to mimic the GNRA-like/receptor interaction L39/H89 from the ribosome. Therefore, we could infer a detailed three-dimensional model for the GUAA/R5.58 interaction (Figure 3), our most abundant receptor interaction which accounted for ∼38% of all the receptor winners (Figure 1C, Supplementary Table S1B). The GUAA/R5.58 module is also proposed to be structurally isosteric to another in vitro selected GUAA/B7.8 module (3), which presents a sequence almost identical to the S8 protein binding site of the 16S rRNA of E. coli (49).
By taking advantage of the present availability of several structural models for different GNRA/receptor interactions, several general principles pertaining to the structural organization of GNRA receptors can be recapitulated. All GNRA receptors take advantage of a type I/II A-minor submodule that involves the 3rd and 4th positions of the GNRA tetraloop interacting with the minor groove side of two Watson–Crick bps, usually two G:C bp. For instance, typical GYRA/helix receptors present a planar type I A-minor between the last adenine of the GYRA and a conserved G:C bp that is associated to a type II A minor interaction between the third position of the GYRA and the Watson–Crick bp located below the conserved type I G:C bp. However, in order to achieve increased binding affinity and GNRA selectivity, a platform module and/or a specific recognition module need to be associated with the A-minor module. This is the case for all GNRA/receptors modules with improved binding affinity versus GYRA/helix modules. For the best GNRA binders, such as the GAAA/11nt and the GGAA/R1 modules, the type I/II A-minor is found tilted (11,33). Likewise, GUAA/S8 and GUAA/R5.58 modules, which have Kds within the same range as those observed for the GAAA/11nt and GGAA/R1 modules, present a tilted type I/II A-minor submodule. This tilted conformation can assure optimal Pi stacking interaction between the second nucleotide position of the GNRA and their respective nucleotide platforms and seems to be more pronounced for the 11nt, R1 and S8-like receptor modules than for IC3-like receptors (Figures 3 and 5B and Supplementary Figure S6). However, the new Ra, Rb and R5c receptors are significantly less selective for their cognate tetraloop than the highly selective GAAA/11nt and the GGAA/R1 modules (11). Many of them have GNRA selectivity comparable to IC3 and Vc2 receptors, although R5 receptors have significantly better GNRA binding affinity than IC3 and Vc2 receptors (Figure 5). Contrasting with the GAAA/11nt and GGAA/R1 receptors (11,33), no apparent specific hydrogen contacts are apparently formed between the second nt position of the GNRA tetraloop and the R5.58, IC3 and Vc2 receptors, preventing high GNRA selectivity. For R5.58, the preference for GUAA versus the other GNRAs might result from steric hindrance with respect of purines and/or from indirect contacts through the formation of a salt bridge as it is observed in the GNRA-like L39/H89 interaction from the ribosome (23). Interestingly, nucleotide changes at positions not directly involved in direct contacts between the GNRA and the receptor can have beneficial synergetic effects on the way the GNRA docks unto the receptor, improving not only the thermodynamics of the tertiary interaction but also its GNRA selectivity. This is observed for the R5b variants with scaffold mutations, the R5c variant 5.15e, and the R5a variants 5.8d and 5.58M28 (See Figure 4 and Supplementary Table S3). Perhaps not so surprising, the thermodynamics and other biophysical properties of RNA tertiary interactions are highly dependent on the structural context within which these interactions take place. Like the 11nt module, 2x_bulge modules likely assemble with their cognate GNRA by an induced fit mechanism. As such, 2x_bulges as well as other RNA modules are not rigid, static building blocks but dynamic modules that adopt preferential conformations within a larger accessible conformational space (50).
Based on our extensive mutational analysis performed on the R5a family and other S8-like receptors (23), 2x_bulge modules have strong mutational robustness. Most 5.58 variants maintain stronger affinity for GNRA tetraloops than GYRA/helix receptors (Figure 2 and Supplementary Figure S7). Besides the A-minor interaction, the main stabilizing contribution comes from Pi stacking interactions between the nucleotide platform submodule and the GNRA tetraloop. Platform modules are likely more tolerant to mutations than specific recognition modules relying on specific H-bonds contacts. Upon some mutations, 2x_bulge receptors can display structural plasticity. Indeed, several variants have been shown to adopt different 2D structure patterns (Supplementary Figure S9) as well as different GNRA selectivity patterns (e.g. Figure 4 and supplementary Figure S11). As described below, these characteristics can shed interesting clues on the structural evolution of RNA at a tertiary structural level.
A large body of studies in RNA molecular evolution have been drawn from computational models linking genotype (sequence) to phenotype as expressed as a secondary (2D) RNA structure (e.g. (51,52). It is only recently that more empirical studies have started to provide insight on the real nature of RNA fitness landscapes (e.g. (53–56)) but to our knowledge, none of them have focused at the level of RNA tertiary folding and assembly level. Herein, we provide a snapshot of the RNA fitness landscape associated to one of the most widespread long-range tertiary modules identified in stable RNA molecules. Our empirical thermodynamic fitness landscape relates the genotype network, including RNA sequence signatures for GNRA receptors, to a phenotype network consisting of tertiary structural modules self-assembling with different GNRA tetraloops. As such, to each RNA receptor signature is associated at least seven different GNRA phenotypes expressed as empirically determined binding energies (Figures 2C, 4 and 5). In a more coarse-grained fashion, the overall phenotype can also be expressed as the ability of a receptor to promote assembly with any GNRA. The genotype network connects the most abundant receptor sequences obtained by in vitro selection (totalizing ∼70 percent of the total number of sequences present in the HGT pool) to natural receptors via single point mutation intermediates. In agreement with computational studies (e.g. (51,52,57)) based on 2D structure phenotypes, the nature of our empirical thermodynamic fitness landscape is rugged, with some receptor variants defining peaks of thermodynamic stability and others valleys (Figures 4 and 5). But, most importantly, it also reveals accessible evolutionary trajectories between the R5a, R5b, R5c, S8 and natural receptors. These mutational pathways are characterized by variations in ΔΔG not exceeding 3 kcal/mol (or variations of Kds ranging between 1 and 4000 nM). The three main characteristics that contribute to evolutionary dynamics within the GNRA/receptor fitness landscape are the mutational robustness of the selected and natural receptors, their evolutionary plasticity, which allows their structure to change upon mutation, and the existence of epistatic effects for mutations in some GNRA/receptor contexts.
The mutational robustness of the R5a, R5b and R5c families can also be understood as phenotypic robustness as most receptor variants maintain tectoRNA bimolecular self-assembly with greater binding affinity than the helical receptors. Very few single point mutations are deleterious for tertiary assembly of R5.58 variants (Figure 2). Interestingly, our RNA fitness landscape illustrates that phenotypic variability and mutational robustness are not opposed to one another. Mutational robustness can also facilitate phenotypic variability. Typically, 2x_bulges receptors recognize several GNRA with decent binding affinity. As such, they are more promiscuous than the more selective 11nt and R1 receptors. While most variants (∼80%) have a preference for GUAA, up to 20% of R5.58 single point mutation variants switch for GGAA/GGGA selectivity. This is somewhat consistent with previous observations on RNA 2D structure phenotypes that suggest that more robust phenotypes have also higher phenotypic variability (57,58). The phenotypic variability of 2x_bulges likely results from their structural plasticity. In fact, 2x_bulge modules have been categorized to belong to at least two different classes: the S8-like and P12-like modules (12,23), which essentially differ in the way their nt platforms are structured. It was proposed that some receptors might be able to conformationally switch from one class to another (23). The evolutionary structural plasticity of 2x_bulge modules is also supported by the observation of epistatic effects taking place upon mutations within different receptors contexts (see Results and Supplementary Figure S11). Additionally, the synergetic effects of scaffold mutations in the R5b family underscore the importance of the context within which long-range tertiary interactions are formed. Mutations far away from the site of interaction can facilitate evolutionary adaptation and can dramatically smooth the GNRA/receptor fitness landscape, enabling the exploration of new phenotypes. Clearly, spreading mutations over a larger RNA structural context confers large scale robustness that dramatically improves fitness (59). However, it remains to be seen whether the type of scaffold mutations associated to GNRA/receptor interactions within the tectoRNA system can translate to other RNA structural contexts.
Perhaps one of the most interesting aspects of the GNRA/receptors landscape is that it illustrates possible evolutionary transitions between the artificial selected receptors R5 and natural receptors (Figure 5). Because of the structural characteristics of 2x_bulge modules, it is readily possible to have these types of modules evolving towards helical receptors in less than two deletion mutations. As shown herein, helical receptors are then able to evolve into the 11nt receptor family via IC3-like receptors (Figure 5). While the transition from R5.58 toward helical receptors leads to a reduction of binding fitness between the receptor and the loop, this type of transition is thermodynamically fluid in the sense that it could easily go both directions. This is somewhat favored by the fact that both categories of receptors recognize best GUAA but still have the potential to recognize other GNRA tetraloops, offering several possible mutational pathways between them. This is also the case for the transition between helical receptors and IC3-like receptors. In contrast, the mutational transition from the IC3-like receptors toward the 11nt receptor is likely more directional. The GAAA/11nt class of receptors is not only thermodynamically favored but also highly selective for GAAA. Because of its lack of structural plasticity for GNRA recognition, the number of energetically favorable mutational pathways between IC3-like and 11 nt-like receptors is dramatically reduced. Within a fitness landscape, the preferential directionality of an evolutionary transition can potentially affect the evolution of some particular molecular features toward unique solutions. In the case of the GNRA/receptor thermodynamic fitness landscape, GAAA/11nt receptors might be informational ‘attractors’ as it might be more difficult to escape the highly favorable GAAA/11nt fitness peak once a GNRA/receptor has evolved into it. In other words, GNRA/receptor modules present within the context of different RNA molecules could have been canalized through evolution toward the GAAA/11nt module into which they were locked in.
Notwithstanding, the strong bias toward GAAA/11nt and GYRA/helix interacting modules observed in natural stable RNAs is not easy to explain based on the sole considerations of thermodynamics, kinetics and loop selectivity. For instance, the thermodynamically weaker GYGA/helix module is almost as widespread as the GAAA/11nt module in many biomolecules (Supplementary Table S5 from (43)), suggesting that other selection pressures resulting from the larger structural context of RNAs, the kinetics constraints on the global folding of RNA inside the cell or the involvement of cellular components are likely at play (60). It is noteworthy that the R5 families of receptors, as well as other GNRA/receptors previously isolated by in vitro selection, have a higher G/C content than the widespread ‘U/A-rich’ GAAA/11nt receptor interaction. While the presence of U/A-rich GNRA/receptor modules might result from the genome enrichment in AT versus GC base pairs observed across organisms, the prevalence of U/A-rich GNRA/receptor modules is also observed at the level of many stable RNA molecules from thermostable organisms. We previously investigated another possible explanation (43) stating that ‘the preferred occurrence of natural RNA module sequences stems from an evolutionary adaptation that makes them less prone to misfolding and therefore less likely to interfere with the folding of a large RNA sequence (through formation of alternative pairings or interactions with other regions of the RNA sequence)’. To this end, several artificial RNA attenuators based on natural and artificial GNRA receptors were tested in vitro for their ability to prevent inter-molecular GNRA/receptor interactions by trapping the receptor sequence into an alternative intra-molecular pseudoknot (PK). Our data indicated that ‘G/C-rich’ receptors were more likely to be trapped into alternative PK structures than ‘A/U-rich’ receptors. We therefore proposed that, in nature, the GAAA/11nt and GYRA/helix interactions result from a negative selection pressure that minimizes kinetic and thermodynamic folding traps in large RNA structural contexts. The first strategy takes advantage of AU-rich internal loop modules, like the 11nt, IC3 and Vc2 receptor motifs, that minimize the formation of stable alternative base pairings. The second strategy takes advantage of receptors like the ‘classic’ GYRA/helix interactions that use Cs and Gs to maximize the formation of stable local Watson–Crick helical regions, preventing the formation of long-range alternative pairings. In other words, when functional RNAs started to grow in size and complexity during the early ‘RNA world’, the sequence pattern of their local constituent structural modules might have been funneled toward unique solutions to minimize sequence interference with the global RNA structural context rather than being funneled towards local energetic minima. Nevertheless, it is still possible that the structural GUAA/R5 modules isolated herein might be identified in RNA molecules from thermostable organisms.
In conclusion, our exploration of the thermodynamic fitness landscape and possible evolutionary trajectories of self-assembling RNA tertiary modules parallels previous studies that investigated the evolutionary trajectories of proteins and that similarly highlighted the importance of epistasis (61). While our selection system restricts the conformational freedom necessary for exploring all possible types of RNA–RNA interactions, it minimizes the isolation of long-range Watson–Crick base pairings that can easily override A-minor mediated interactions during SELEX because of their greater thermodynamic stability (e.g (11)). As such, our present system is interesting for exhaustively exploring novel A-minor mediated tertiary modules targeting at other RNA loops. Alternatively, selection systems taking advantage of other structural contexts like the DSL ligase (30,62), might provide additional clues on the range and uniqueness of A-minor mediated GNRA-receptor modules reachable by RNA. It is also worth mentioning that our present study expands the tool box of RNA modules for RNA nanotechnology and RNA synthetic biology (e.g. (63–66)). Our various selective tetraloop-receptor modules could be used as building blocks to enable greater regioselective control in nano-construction and derive novel programmable RNA architectures of increasing structural and functional complexity (e.g. (35,67–70)).
ACKNOWLEDGEMENTS
The authors thank Maria del Carmen Jaeger for fruitful discussions, Ryan Cohen for helping with Levenshtein distance matrices, and Celia Blanco and Evan Jansen for helping with high throughput sequence analysis. This work is dedicated to the memory of Prof. Albert Jaeger, MD and Prof. Jérome Lejeune, MD.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Aeronautics and Space Administration (NASA) [80NSC17K0031 to L.J., in part]; UCSB academic senate, intramural research grants (to L.J.). Funding for open access charge: UCSB Open Access Fund.
Conflict of interest statement. None declared.
Notes
Present address: Paul Zakrevsky, RNA Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA.
Present address: Erin R. Calkins, Department of Biological Sciences EBS 212, Santa Barbara City College, 721 Cliff Dr., Santa Barbara, CA 93109, USA.
Present address: Vasken L. Keleshian, UC Irvine medical center, 333 City Blvd. West, Suite 400, C3, 4080 Orange, CA 92868, USA.
Present address: Stephanie Baudrey, Architecture et Réactivité de l’ARN, CNRS, Université de Strasbourg, UPR 9002, F-67000 Strasbourg, France.
REFERENCES
1.
2.
3.
4.
5.
6.
7.
8.
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
24.
25.
26.
27.
28.
29.
30.
31.
32.
33.
34.
35.
36.
37.
38.
39.
40.
41.
42.
43.
44.
45.
46.
47.
48.
49.
50.
51.
53.
54.
55.
56.
57.
59.
60.
61.
62.
63.
65.
66.
67.
68.
69.
70.
 In vitro selected GUAA tetraloop-binding receptors with structural plasticity and evolvability towards natural RNA structural modules
In vitro selected GUAA tetraloop-binding receptors with structural plasticity and evolvability towards natural RNA structural modules
