
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
- Altmetric
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides with little or no protein coding potential. The expanding list of lncRNAs and accumulating evidence of their functions in plants have necessitated the creation of a comprehensive database for lncRNA research. However, currently available plant lncRNA databases have some deficiencies, including the lack of lncRNA data from some model plants, uneven annotation standards, a lack of visualization for expression patterns, and the absence of epigenetic information. To overcome these problems, we upgraded our Plant Long noncoding RNA Database (PLncDB, http://plncdb.tobaccodb.org/), which was based on a uniform annotation pipeline. PLncDB V2.0 currently contains 1 246 372 lncRNAs for 80 plant species based on 13 834 RNA-Seq datasets, integrating lncRNA information from four other resources including EVLncRNAs, RNAcentral and etc. Expression patterns and epigenetic signals can be visualized using multiple tools (JBrowse, eFP Browser and EPexplorer). Targets and regulatory networks for lncRNAs are also provided for function exploration. In addition, PLncDB V2.0 is hierarchical and user-friendly and has five built-in search engines. We believe PLncDB V2.0 is useful for the plant lncRNA community and data mining studies and provides a comprehensive resource for data-driven lncRNA research in plants.
INTRODUCTION
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides with little or no protein coding potential (1–4). They account for a large portion of transcripts in cells; for instance, there are 189 901 lncRNAs annotated in RNAcentral (5) for human compared with 19 288 protein-coding genes in ENSEMBL (6). Mammalian lncRNAs have been shown to regulate gene expression and other cellular processes and are implicated in cancer and other diseases (7). In addition to functioning as miRNA sponges (8), plant lncRNAs are also known to interact with regulatory proteins to modulate gene expression (1,3,9–11). From the earliest investigations of important lncRNAs including COLDAIR/COOLAIR (12) and IPS1 (13) almost two decades ago, recent years have seen rapidly growing interest in the functional exploration of individual plant lncRNA (14,15).
The expanding list of lncRNAs and accumulating functional evidence in plants have necessitated the development of a comprehensive database to act as a data repository and a platform for lncRNA analysis. Multiple databases have thus been developed in the past several years (16), such as lncRNAdb (17), NONCODE (18), EVLncRNAs (19), PLNlncRbase (20), CANTATAdb (21), GreeNC (22), RNAcentral (5) and PLncDB V1.0 (1), all of which are devoted to archiving plant-related lncRNA information (Supplementary Tables S1 and S2). These databases have greatly facilitated lncRNA studies, but they may have some deficiencies, such as uneven annotation standards, and they lack of some of the important features for lncRNAs, including expression information, potential target information and epigenetic signals. For instance, RNAcentral (5), which attempted to collect lncRNAs from several important databases, lacks standard criteria for lncRNA annotation. Moreover, it only contains basic lncRNA features, such as genome coordinates and sequence information, without expression or target information. EVLncRNAs (19), lncRNAdb (17) and PLNlncRbase (20) mainly focused on experimental validated lncRNA candidates, which resulted in limited number of lncRNAs in these databases. As the most comprehensive databases for plant lncRNA research, although CANTATAdb (21) and GreeNC (22) attempted to annotate plant lncRNAs with standard criteria, the number of lncRNAs and annotated plant species is still far from sufficient due to the limited datasets they used. Moreover, these two databases are incompatible with known validated lncRNA information, for example, the well-known lncRNA COLDAIR cannot be found either by keyword search or by sequence blast. Thus, it is highly desirable to establish a plant lncRNA database with more uniform and strict annotation standards, greater data integrity and more comprehensive annotations.
Since its launch, PLncDB has attracted wide interest from the plant lncRNA research community. As of August 2014, PLncDB was inducted into RNAcentral as a third party data specialist database. In the present study, with the goal of creating a one stop plant lncRNA database, we upgraded PLncDB V2.0 to use a standard annotation criteria to systematically annotate 1 246 372 lncRNAs in 80 phylogenetically representative plant species. Known and experimentally verified lncRNAs were collected from EVLncRNAs, and plant lncRNA candidates in RNAcentral were also integrated into our database. PLncDB V2.0 also provides an easy-to-use interface to browse, search, and download data, enabled by several search engines. In addition, PLncDB V2.0 deploys several tools (JBrowse (23), eFP Browser (24), EPexplorer and so on) to visualize expression patterns and epigenetic signals for lncRNAs. Taken together, PLncDB V2.0 is a comprehensive functional database amenable for data mining and database-driven research therefore providing a useful resource for the plant lncRNA research community.
MATERIALS AND METHODS
Data sources
The current version of PLncDB (version 2.0) consists of lncRNA entries from 80 plants from chlorophytes to embryophyta, including 16 monocotyledons and 53 eudicotyledons (Figure 1A, Supplementary Table S3). Whole genome references and gene annotations were downloaded from NCBI (25), Phytozome V12.1 (26), and other official websites for model plants (Arabidopsis: TAIR; Tobacco/tomato/potato: Sol Genomics Network (27); Maize: MaizeGDB (28); Rice: MSU RGAP (29)). More detailed information is in Supplementary Table S3. In total, 13 834 RNA-Seq datasets were obtained from the NCBI SRA Database (https://www.ncbi.nlm.nih.gov/sra/); details are in Supplementary Table S4. For model plants, we specially collected datasets from studies focusing on transcriptome landscape by mining literatures. Meanwhile, we downloaded RNA-Seq datasets from NCBI for each plant searching by scientific names. The RNA-Seq datasets missing information about tissue, development stage/age, or treatment were discarded. In addition, methylation, histone and small RNA sequencing data were also collected for model plants (Supplementary Table S5). The known experimentally verified lncRNAs were collected from EVLncRNAs, and lncRNA candidates of plants in RNAcentral were also integrated into our database. For model plants, we also collected several well-studied lncRNAs from other studies (3,4,30).

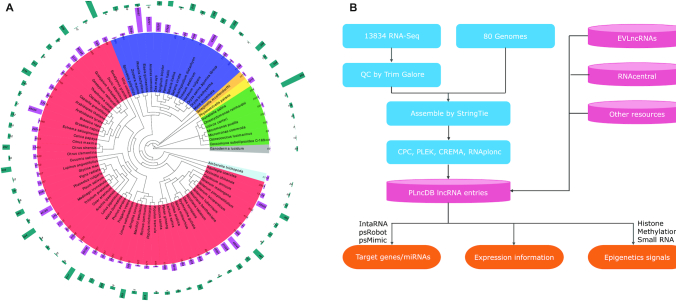
Phylogenetic tree of 80 species included in our analysis and data processing workflow of PLncDB V2.0. (A) Phylogenetic tree of 80 species in PLncDB V2.0. From the inside to the outside: phylogenetic tree of 80 species; numbers of identified lncRNAs and numbers of RNA-Seq datasets used in the analysis pipeline. (B) Data processing workflow and outcomes of PLncDB V2.0.
Data analysis pipelines
Data pre-processing
Linux version 2.8 of the SRA toolkit was employed to convert original compressed sra files into Fastq format. Trim Galore (version 0.50) (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) was used to trim adapter sequences with parameters ‘-q 30 –length 35’ (Figure 1B).
Annotation of lncRNAs
Clean data were aligned to the reference genome using the read aligner HIAST2 (31). The transcriptome of each library was assembled separately using StringTie (32), and all gtf result files were then merged into one for each species with StringTie –merge (Figure 1B). Then, we compared the assembled transcript isoforms with the reference genome annotation information, which represents all protein coding gene models identified in each plant genome. Transcripts with a length shorter than 200nt and an open reading frame (ORF) length longer than 120 amino acids were discarded (https://www.ncbi.nlm.nih.gov/orffinder/). In order to remove transcripts that may encode short peptides, the Swiss-Prot database was searched using the blastx program with the parameters -e 1.0e-4 -S 1. Further, the transcripts overlapping with rRNA, tRNA, sRNA and miRNA in Rfam database were filtered. The CPC (33), PLEK (34), RNAplonc (35) and CREMA (36) programs were used to calculate the coding potential of the remaining transcripts. Only transcripts satisfied with at least two criteria were considered as lncRNA candidates in our database. High-confidence candidates were defined if more than three criteria were satisfied, and the left were defined as medium-confidence candidates.
Expression analysis
The expression values for lncRNAs were normalized by TPM as previously described (37). For each RNA-Seq dataset, reads were mapped to the reference genome by HIAST2 (31), and TPMs were calculated using StringTie (32). The tissue specific expressed lncRNA candidates were measured by ROKU (38) with cutoff 0.8.
Target prediction
The regulatory relationship between miRNAs and protein coding genes were predicted by psRobot (39). Mimic target prediction between lncRNAs and miRNAs was conducted by psMimic (40). The interactions between protein coding genes and lncRNAs were predicted by IntaRNA (41) based on secondary structures.
Database development
PLncDB V2.0 was constructed using the Python language (https://www.python.org/), Vue.js (https://vuejs.org/), ElementUI (https://element.eleme.io/#/), and Django (https://www.djangoproject.com/) framework. Network proxy services were provided through nginx (https://www.nginx.com/). The graph and network were visualized using Echarts (https://echarts.apache.org/zh/index.html) and Cytoscape (42).
RESULTS
Data content of PLncDB V2.0
Data in the current version of PLncDB (version 2.0) were based on 13 834 RNA-Seq datasets from 80 plant species ranging from chlorophytes to angiosperms (Figure 1A and Supplementary Table S3). Using a standard method with strict annotation criteria (Figure 1B), 1 246 372 lncRNA transcripts were discovered. Moreover, experimentally verified lncRNAs from EVLncRNAs and plant lncRNAs collected in RNAcentral were also integrated into PLncDB. Due to the difficulty of combining lncRNAs with overlapped regions from different sources, PLncDB currently considers these candidates independently and provides the corresponding source information for users. In addition, PLncDB provides information on epigenetic modification signals on lncRNA-encoding loci and their flanking genomic regions by analyzing epigenetic sequencing data. Compared with other plant lncRNA databases, the coverage of lncRNA annotation was greatly increased in PLncDB V2.0 (Figure 1A, Supplementary Tables S1 and S2, Supplementary Figure S1). In addition, PLncDB V2.0 provides expression patterns for lncRNAs across organ types and developmental stages for all species and potential targets were also predicted by IntaRNA (41) and psMimic (40).
lncRNA features of PLncDB V2.0
Similar to other databases, PLncDB includes all basic information of a given lncRNA, including nucleotide sequence and longest ORF sequence, predicted secondary structure and genome coordinates. Moreover, lncRNA annotation in PLncDB has several unique features. First, since all annotated lncRNAs were predicted from RNA-Seq datasets, lncRNA expression patterns were directly obtained from normalized TPM of these RNA-Seq datasets. Hence, expression of these lncRNAs can be directly used to cluster lncRNA candidates with similar functions (Figure 2A–C). Using heatmaps, we could easily identify some lncRNAs with tissue-specific expression (Figure 2A). Second, except for the known targets of several lncRNAs, the prediction of other targets in PLncDB V2.0 was done using psRobot (39), psMimic (40) and IntaRNA (41) (Figure 2D). Hence, users can predict the putative functions for lncRNAs by their potential targets. Third, expression of thousands of lncRNAs is associated with epigenetic regulation. Yet, little is known about epigenetic regulation of lncRNA expression itself in plants. Hence, in PLncDB V2.0, we analyzed ChIP-chip/ChIP-Seq data of the following histone modifications [H3K27me3, H3K4me3, H3K36me3 and H3K9me3] (Figure 2B). Moreover, we profiled expression changes of lncRNAs in multiple RdRM-related mutants [RDD, DCL1/2/3/4, AGO4, RDR2 and DMS1].

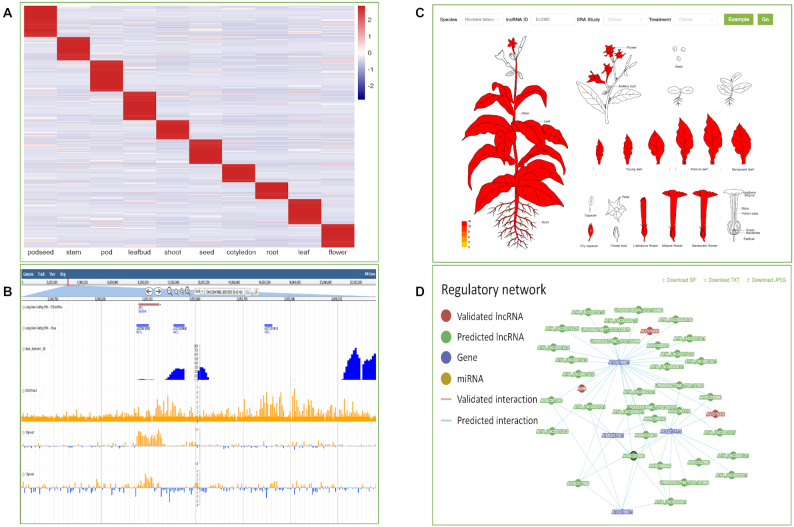
Unique features of PLncDB V2.0. (A) An example of lncRNA expression patterns by EPexplorer. Expression values (TPM) of all lncRNAs in soybean were obtained from different tissues including leaf, root, seed, stem, etc (B) Visualization of expression patterns and epigenetic modification for lncRNAs (IPS1) by JBrowse. (C) Visualization of expression patterns for one lncRNA in tobacco using eFP Browser. (D) Example of validated and predicted lncRNA targets for one lncRNA in Arabidopsis.
Functions of PLncDB V2.0
PLncDB V2.0 provides convenient access, functional search engines and powerful analytical tools (Figure 3). Users can browse all data by shortcuts and multiple layers of webpages. All data can be downloaded in FTP bulk and customized manners (Figure 3).


A schematic view of PLncDB V2.0 features.
Browse
Users can browse lncRNAs using species shortcuts on the home page or through the Browse tag in the toolbar. The Browse tag will guide user to the species lists where the species phylogenetic information is included. Upon clicking the species name, the summary information including the reference genome version, the numbers of predicted and validated lncRNAs and the number of RNA-Seq datasets will be displayed. The next level of Browse is a summary of all lncRNAs and genes in a specific species where users can check detailed information. Further, when clicking each lncRNA candidate, the link will direct users to a detailed information page, consisting of the following six sections: basic type information, sequence information, secondary structure, genomic map information, regulatory network and expression pattern information. For validated lncRNA candidates, it also contains the corresponding experimental information extracted from EVLncRNAs, and the related PubMed literature.
Search
Users can search the whole database with five search engines. Using keyword or lncRNA identifier search engine located on the webpage banner, users can search all fields of the whole database, and the result will be a summarized of a list of all hit lncRNAs. The second search engine is by sequence, and here a BLAST web interface is deployed. For the convenience of quickly searching for lncRNA-related information such as genome coordinates and expression values, PLncDB V2.0 provides the respective search engines. Meanwhile, PLncDB also supports tissue specific expressed lncRNA candidates search for convenience. All the search results can be downloaded in FASTA, CSV and GFF format.
Tools
We have constructed three useful tools including EPexplorer (Figure 2A), JBrowse (Figure 2B) and eFP Browser (Figure 2C) for PLncDB V2.0. By EPexplorer, we could obtain expression landscape for batch lncRNA candidates and identify tissue specific expressed candidates easily (Figure 2A). The online genome browser with a large set of transcriptome data will be useful for biologists to further investigate functional roles of lncRNAs. With the eFP Browser (Figure 2C), users can quickly obtain the expression landscape for a specific lncRNA, and then design appropriate experiments to verify their hypothesis. In our study, we analyzed Chip-chip and Chip-Seq data for histone modifications including H3K27me3, H3K4me3, H3K36me3 and H3K9me3 and so on (Figure 2B). We also profiled expression changes for some lncRNAs in the context of several RdRM-related mutants, such as RDD, DCL1/2/3/4, AGO and RDR2. These results can be easily accessed using JBrowse for each plant. For instance, for IPS1 in Arabidopsis, using JBrowse (Figure 2B), we can easily find that IPS1 is expressed in response to phosphate starvation. In addition, there are some epigenetic signals (H3K27me3) around its flanking region. For enod40 candidate of tobacco, we find that in addition to expression in stem and leaf (Figure 2C), this lncRNA is also expressed in flower tissue, implying that it may also regulate flower development.
Download
Through the Download tag located in the toolbar, all basic information on lncRNAs in each species could be downloaded in bulk or for specific species. Users can download data using three ports on the Download page. First, by FTP link, users can browse all available information in bulk. Second, via the port of user-defined Download, users can choose the information they are interested in by selecting species and data types. Third, quick access is provided to users so that the genome coordinates of lncRNAs could be readily downloaded. Meanwhile, a backup copy of all data in PLncDB was also uploaded to Zenodo (https://zenodo.org/) with ID of 4017591.
Submit
In an effort to make PLncDB a one stop community resource, we have begun to accept submissions of plant lncRNAs with required information alone or in batch. All submitted lncRNA items will be processed by our standard procedure described in the Material and Methods. By this process, the submitted lncRNAs will be grouped into categories.
DISCUSSION
In the past two decades, great efforts have been made toward the identification, characterization and functional analysis of plant lncRNAs. In addition, abundant data for RNA-Seq and Chip-Seq have accumulated as a result of rapid development of next generation sequencing techniques. Together, they have provided a solid foundation for annotating plant lncRNAs and, therefore, there is an urgent need to construct a comprehensive database to store and organize all knowledge relating to plant lncRNAs. Currently, CANTATAdb (21) and GreeNC (22) are the most comprehensive databases for plant lncRNA research, but the number of lncRNAs and annotated plant species is still far from sufficient due to the limited datasets they used. EVLncRNAs mainly focused on validated lncRNA candidates thereby limiting its wide-spread usage. Therefore, we set out to use a standardized pipeline with the latest plant lncRNA annotation criteria to annotate lncRNAs for all plant species with genome references and available RNA-Seq datasets (Figure 1B and Supplementary Table S4).
Compared to CANTATAdb/GreeNC/EVLncRNAs and other lncRNA databases, not only more plant species were annotated, but the RNA-Seq datasets and epigenetic information have also been improved in PLncDB V2.0 (Supplementary Tables S1 and S2). As shown in Figure 1, the number of annotated lncRNAs was improved substantially. One of the immediate consequences of this improvement is that users can now conduct comparative analysis of lncRNAs among different tissues and species (Figure 2).
In addition, several features for plant lncRNAs were provided in PLncDB V2.0. By normalizing expression levels across different tissues, development stages, or stress treatments, lncRNA expression patterns in species can be easily established (Figure 2A and C). Analysis of the epigenetic dataset clearly showed that lncRNA genes are regulated by methylation and histone modification (Figure 2B). Target collection and prediction provides information on the regulatory network for specific lncRNA and increases our understanding of plant lncRNAs based on available information (Figure 2D). From the tissue specific expression analysis function provided in PLncDB V2.0, we could find a lot of lncRNAs with tissue specific expression pattern for most of plant species (Figure 2A), which is consistent with other findings (43–46). Compared to protein coding genes, the tissue specific expression of lncRNAs provides important clues about their specific functions within tissues (2).
Meanwhile, PLncDB V2.0 provides a user-friendly interface to browse and access all data via multi-layer webpages, powerful search engines and download ports (Figure 3). At the same time, we plan to update and improve PLncDB semi-annually, by integrating data from other databases and researchers. For example, there are still some plants not covered by PLncDB V2.0 due to the huge amount of computation needed, which will be integrated in the next update. We will use the same standard pipeline for annotating new entries and add them into PLncDB. Hence, we believe PLncDB V2.0 will be a comprehensive functional database for data mining and database-driven research, and therefore a useful platform for the plant lncRNA community.
DATA AVAILABILITY
PLncDB V2.0 is freely available at http://plncdb.tobaccodb.org/.
ACKNOWLEDGEMENTS
We thank Mr. Zeqing Guo, Keqiang Hu and Yongsheng Yan for their IT support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Zhengzhou Tobacco Research Institute [CNTC: 110201901024(SJ-03), 110202001020(JY-03), 110201601033(JY-07)]; China Association for Science and Technology [Young Elite Scientists Sponsorship Program 2016QNRC001]; National Research Foundation of Singapore [RSSS grant: no. NRF-RSSS-002 to N.H.C]. Funding for open access charge: China Association for Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
1.
2.
3.
4.
5.
6.
8.
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
24.
25.
26.
27.
28.
29.
30.
31.
32.
33.
34.
35.
36.
37.
38.
39.
40.
41.
42.
43.
44.
45.
 PLncDB V2.0: a comprehensive encyclopedia of plant long noncoding RNAs
PLncDB V2.0: a comprehensive encyclopedia of plant long noncoding RNAs
