
The authors wish it to be known that, in their opinion, the first three authors should be regarded as joint First Authors.
- Altmetric
An updated Lnc2Cancer 3.0 (http://www.bio-bigdata.net/lnc2cancer or http://bio-bigdata.hrbmu.edu.cn/lnc2cancer) database, which includes comprehensive data on experimentally supported long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) associated with human cancers. In addition, web tools for analyzing lncRNA expression by high-throughput RNA sequencing (RNA-seq) and single-cell RNA-seq (scRNA-seq) are described. Lnc2Cancer 3.0 was updated with several new features, including (i) Increased cancer-associated lncRNA entries over the previous version. The current release includes 9254 lncRNA-cancer associations, with 2659 lncRNAs and 216 cancer subtypes. (ii) Newly adding 1049 experimentally supported circRNA-cancer associations, with 743 circRNAs and 70 cancer subtypes. (iii) Experimentally supported regulatory mechanisms of cancer-related lncRNAs and circRNAs, involving microRNAs, transcription factors (TF), genetic variants, methylation and enhancers were included. (iv) Appending experimentally supported biological functions of cancer-related lncRNAs and circRNAs including cell growth, apoptosis, autophagy, epithelial mesenchymal transformation (EMT), immunity and coding ability. (v) Experimentally supported clinical relevance of cancer-related lncRNAs and circRNAs in metastasis, recurrence, circulation, drug resistance, and prognosis was included. Additionally, two flexible online tools, including RNA-seq and scRNA-seq web tools, were developed to enable fast and customizable analysis and visualization of lncRNAs in cancers. Lnc2Cancer 3.0 is a valuable resource for elucidating the associations between lncRNA, circRNA and cancer.
INTRODUCTION
Cancer is a major public health problem and a leading cause of morbidity and mortality worldwide (1). Over the past decades, new cellular roles of RNA molecules have been discovered. It has been established that the dynamics of gene expression in cancer pathology are remarkably complex (2). Long non-coding RNAs (lncRNAs) have attracted increasing attention as cancer biomarkers for early screening, diagnosis, prognosis and analysis of treatment responses (3,4). Circular RNA (circRNA) is a new member of the non-coding cancer genome and exhibits distinct properties and diverse cellular functions (5,6). To facilitate the study of cancer-associated lncRNAs, we previously reported the first and second version of the Lnc2Cancer database (Lnc2Cancer 1.0 and 2.0), which enabled users to search all known experimentally supported lncRNAs associated with various human cancers (7,8).
The growing interest in human lncRNAs and circRNAs as well as the availability of high-throughput and single-cell technologies has resulted in a rapid increase in the number of cancer-related lncRNAs and circRNAs. Notably, cancer-associated lncRNAs and circRNAs can be divided into different groups based on their regulatory mechanisms, biological functions or clinical application. The roles of lncRNAs and circRNAs in cancer-related regulatory mechanisms involving enhancers (9,10), genetic variants (11,12), microRNA (miRNA) interactions (13,14), transcription factors (TFs) (15) and modifications by methylation (16) have also been widely studied. Additionally, novel biological functions of cancer-related lncRNAs and circRNAs have emerged. In recent years, increasing evidence has suggested that lncRNAs and circRNAs play important functions in the cell growth (17,18), apoptosis (19,20), autophagy (21), epithelial mesenchymal transformation (EMT) (22), immunity (23,24) and coding ability (25). Moreover, it has been proposed that lncRNAs and circRNAs could serve as non-invasive biomarkers for exosome circulation (26), drug resistance (27,28), cancer prognosis (29), metastasis (30), and recurrence (31). However, no specialized resource has been devoted to collecting, storing, and distributing these data.
High-throughput RNA sequencing (RNA-seq) has emerged as a powerful method for transcriptomic analysis. It is widely employed for investigating the functions and biological patterns of lncRNAs, finding candidate drug targets, and identifying biomarkers for cancer classification and diagnosis (32). Recent advances in the technologies involving tissue dissociation and high-throughput sequencing at a single cell level have enabled the generation of single cell RNA sequencing (scRNA-seq) datasets, which have been increasingly deposited into the public domain (33). Large amounts of scRNA-seq and RNA-seq data have created new opportunities for data mining and deeper understanding of the lncRNA functions. The development of fast and customizable lncRNA analysis and visualization methods is a key problem in cancer research. Convenient and efficient web tools could fill the gap between cancer-associated lncRNA data and the delivery of integrated information to the end users, thus utilizing the current lncRNA data resources to study human cancers.
To achieve this, we have updated the Lnc2Cancer database to version 3.0 (Lnc2Cancer 3.0) (Figures 1, 2 and Table 1). The current version of Lnc2Cancer documents 10 303 entries of associations between 2659 human lncRNAs, 743 circRNAs, and 216 cancer subtypes. This was achieved by a comprehensive review of >15 000 published papers. Lnc2Cancer 3.0 provides information on experimentally supported regulatory mechanisms (miRNA, TF, genetic variant, methylation and enhancer), biological functions (cell growth, apoptosis, autophagy, EMT, immunity and coding ability) as well as clinical applications (metastasis, recurrence, circulation, drug-resistance, and prognosis) of lncRNAs in human cancer. Two interactive web tool platforms including RNA and scRNA-seq expression data were developed to allow exploration of the involvement of lncRNAs in cancers using a standard processing pipeline. We expect the Lnc2Cancer 3.0 will serve as an important resource for researchers studying the relations between lncRNA, circRNA, and cancer. All information about Lnc2Cancer 3.0 is freely available at http://www.bio-bigdata.net/lnc2cancer or http://bio-bigdata.hrbmu.edu.cn/lnc2cancer.

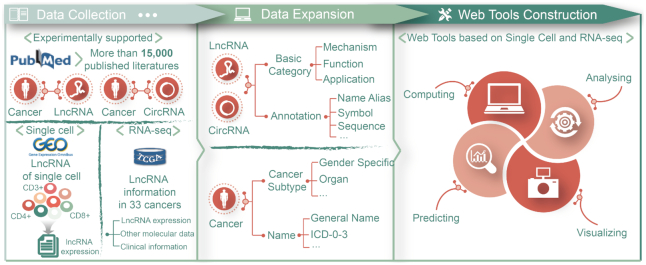
Content of Lnc2Cancer 3.0. The figure summarizes the content of the database, which includes a collection of previously reported cancer-related lncRNAs and circRNAs, comprehensive cancer data, information on lncRNAs and circRNAs, and the construction of single cell and RNA-seq web tools.

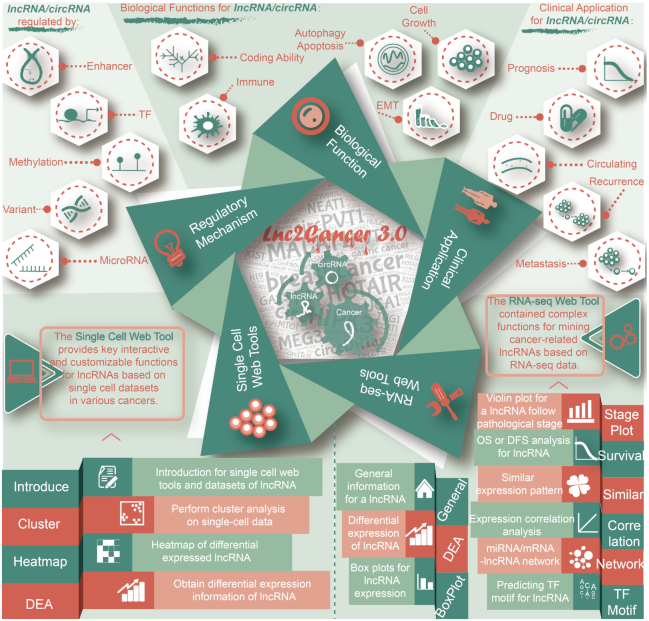
Interface of Lnc2Cancer 3.0. Data on the regulatory mechanisms, biological functions, and clinical applications of lncRNAs and circRNAs in cancers are included. A panel of tools has been developed to mine, visualize and analyze lncRNAs at single cell and RNA-seq levels.

| Features | Lnc2Cancer 2.0 | Lnc2Cancer 3.0 | Fold increase |
|---|---|---|---|
| LncRNA-cancer associations | 4989 | 9254 | 1.85 |
| CircRNA-cancer associations | - | 1049 | New |
| Cancer subtypes | 165 | 216 | 1.31 |
| LncRNAs | 1613 | 2659 | 1.65 |
| circRNAs | - | 743 | New |
| Regulatory mechanisms of LncRNAs, circRNAs | 1894, - | 4076, 726 | 2.15, New |
| Biological functions of LncRNAs, circRNAs | -, - | 4476, 685 | New, New |
| Clinical applications of LncRNAs, circRNAs | 2887, - | 6364, 695 | 2.20, New |
| Single cell Web Tools | - | 49 datasets | New |
| RNA seq Web Tools | - | 33 datasets | New |
IMPROVED EXPANSION AND NEW FEATURES
Data expansion and pre-processing
Lnc2Cancer 3.0 was updated to contain an increased number of associations between lncRNAs or circRNAs and different cancer subtypes (Figure 1 and Table 1). In the first instance, we screened >8000 studies in the PubMed database. Among them, 6500 reports from 2018 to 2020 concerned lncRNAs and 1570 publications from 2017 to 2020 involved circRNAs (34). All searches followed similar keyword combinations as the ones used in Lnc2Cancer 2.0. In addition, to obtain more detailed data, we re-screened >6500 studies in the PubMed database (predominantly reports published before 2015), which had been included in Lnc2Cancer 2.0.
We subsequently extracted experimentally supported lncRNA or circRNA cancer associations, which were confirmed by strong experimental evidence, including RNA interference (RNAi), in vitro knockdown, western blot, real-time quantitative polymerase chain reaction (qRT-PCR) or luciferase reporter assay. If the regulatory mechanisms, biological functions and clinical applications of lncRNA and circRNA were verified, the information was extracted. Additionally, some high-quality scRNA-seq expression data of lncRNAs in cancers were extracted. These included different cancer subtypes, cell numbers, lncRNA numbers, cell lines and tissues. Concurrently, detailed information on lncRNA, circRNA and cancer was recorded. The methods and principles of collecting the data can be found in Lnc2Cancer 2.0.
Furthermore, we added more detailed data to demonstrate the lncRNA or circRNA cancer associations more comprehensively. Other lncRNA nomenclature, including aliases, synonyms, gene ID, names from HGNC (35), Ensembl (36), and GENCODE (37) as well as Genbank (38) and Refseq ID, was collected. In addition, Arabic-coded (circBase) (39) and host gene names (HUGO ID from Circbank) (40) were included in the database. These names were used to combine the synonyms for lncRNAs and circRNAs to ensure that the information for coincident lncRNAs and circRNAs was the same. A standardized classification scheme, namely the International Classification of Diseases for Oncology 3rd Edition (ICD-O-3), was employed to annotate each cancer type. The above data expansion and pre-processing involved a systematic review of 15 000 published papers. The current version of Lnc2Cancer includes 8297 entries of associations between 2775 human lncRNAs and 220 human cancer subtypes. Lnc2Cancer 3.0 also contains 1049 entries of associations between 743 human circRNAs and 64 human cancer subtypes.
Experimentally supported regulatory mechanisms, biological functions and clinical applications of lncRNAs and circRNAs in cancer
To comprehensively characterize the roles of lncRNAs and circRNAs in cancer, we manually curated their regulatory mechanisms, biological functions, and clinical applications. The information we collected had to be verified by high-quality experiments. Conservation of N6-methyladenosine (m6A) and peptide was analyzed to determine the lncRNA coding ability. If available, the types of immune cells involving immune-related lncRNAs and circRNAs in cancer were also extracted. Only the expression of lncRNAs or circRNAs in the blood, exosome, plasma or serum was defined as circulating lncRNAs or circRNAs. Additionally, those defined as circulating RNAs in the evaluated studies were included. Predominantly, lncRNAs and circRNAs detected in exosomes were marked. An alternative detailed method and principle of collecting data can be found in Lnc2Cancer 2.0. Overall, Lnc2Cancer 3.0 provides a systematic pipeline including regulatory mechanisms, biological functions, and clinical applications of lncRNAs and circRNAs in cancer.
Newly developed single cell web tool for discovery and analysis lncRNA
As a result of rapid expansion of the available expression profiles obtained by high-throughput sequencing technologies at a single cell level, the development of an efficient approach for the analysis of large amounts of datasets is essential. A rapid and comprehensive method would enable the analysis of cancer pathology and discovery of lncRNAs as cancer biomarkers. In Lnc2Cancer 3.0, we designed a single cell web tool, which can be employed to identify novel cancer-related lncRNAs according to the provided single cell datasets. Forty-nine single cell datasets concerning lncRNA expression, including 20 cancer types and 22 100 cells, were collected from Gene Expression Omnibus (GEO: https://www.ncbi.nlm.nih.gov/geo/). The single cell web tool is equipped with three key functions. The Cluster function allows users to perform cluster analysis of single cell lncRNA expression data based on UMAP and t-SNE dimensionality reduction methods. Moreover, the Heatmap function provides a heatmap of differentially expressed lncRNAs among diverse clusters. Lastly, the Differential Expression Analysis (DEA) function enables users to obtain differential expression information and violin plots of lncRNAs. All of the above functions can be performed using the R package Seurat (version 3.1.5).
Newly developed RNA-seq web tool for mining and deeper understanding of cancer-related lncRNAs
Recent technical advances in large-scale sequencing and genomics methods have provided a valuable platform for mining novel cancer-related lncRNA biomarkers as well as for investigating biological functions of lncRNAs. In this work, we obtained RNA-seq datasets containing information of lncRNA expression. In total, 15 878 lncRNAs, 33 cancer types, 9664 tumors and 711 normal control samples were identified from The Cancer Genome Atlas (TCGA: https://portal.gdc.cancer.gov). Nine functions of the RNA-seq tool were established: (i) the general function allows users to construct crosstalk between low- and high-throughput experiments involving cancer-related lncRNAs. It provides general information on the subcellular localization (from lncATLAS) (41), functions (from LncBook) (42), gene ontology annotation (from LncBook), mean expression in cancer and normal tissues, and box plots for a specific lncRNA in various cancers; (ii) the DEA function enables users to obtain differential lncRNA expression analysis and heatmaps utilizing diverse custom statistical methods and thresholds for specific cancers; (iii) the Boxplot function generates box plots with custom colors for comparing the expression of a specific lncRNA in cancer and normal samples; (iv) the Stage Plot function produces expression violin plots for specific lncRNAs based on major and detailed pathological stage; (v) the Survival function performs overall survival (OS) or disease free survival (DFS, also called relapse-free survival [RFS]) analysis based on median and quantile expression values of specific lncRNAs; (vi) the Similar function identifies a list of lncRNAs with similar expression patterns using an input lncRNA and a selected cancer type; (vii) the Correlation function provides lncRNA expression correlation analysis based on custom methods, including Pearson, Spearman, and Kendall, for two cancer-related lncRNAs; (viii) the Network function gives information on the miRNA–lncRNA and mRNA–lncRNA co-expression networks; (ix) the TF motif function predicts a TF motif for a specific lncRNA and provides a TF motif sequence LOGO figure.
DATABASE CONSTRUCTION AND IMPROVED USER INTERFACE
All data in Lnc2Cancer 3.0 were stored and managed using the MySQL (version 5.7.18) data server. The web interfaces were built in JSP on Linux and Apache platforms. Lnc2Cancer 3.0 is freely available at http://www.bio-bigdata.net/lnc2cancer and http://www.bio-bigdata.com/lnc2cancer. The old versions, namely Lnc2Cancer 1.0 and 2.0, also remain available. Users can access them either from the Lnc2Cancer 3.0 homepage or directly at http://bio-bigdata.hrbmu.edu.cn/lnc2cancer1.0/ and http://www.bio-bigdata.net/lnc2cancer2.0/.
Lnc2Cancer 3.0 exhibits a user-friendly interface and provides flexible routes for data access, enabling users to query the database in just a few steps. (i) From the ‘Browse’ page, users can browse all experimentally supported associations of lncRNAs, circRNAs and primary cancer tissues (Figure 3A). In the cancer-Centric section, there are two ways to search the data. The first includes anatomical classification using a human bodymap, while the second is a list of cancer types. In the lncRNA and circRNA-Centric section, users can browse by diverse regulatory mechanisms, biological functions, and clinical applications of lncRNAs and circRNAs. (ii) The ‘Search’ page provides ‘general search’ and ‘advanced search’ options (Figure 3B–D). Using the general search, users can search the database by lncRNA and cancer names. In the advanced search, users can input more detailed and systemic information by restricting the outputs based on deregulated expression patterns, samples, RNA type, regulatory mechanisms, biological functions, and clinical applications. (iii) From the ‘Single Cell Web Tool’ page, users can utilize interactive and customizable functions, including accessing general information, clustering, heatmap generation and differential expression analysis for lncRNAs based on 49 single cell datasets (Figure 4A). (iv) From the ‘RNA-seq web tool’ page, users can perform complex functions and obtain detailed data on cancer-related lncRNAs, including general information, differential expression analysis, box plotting, stage plotting, survival analysis, similar lncRNAs identification, correlation analysis, network construction and TF motif prediction (Figure 4B). (v) Lnc2Cancer 3.0 is a completely open resource, meaning that users can obtain all data from the ‘Download’ page. (vi) From the ‘Help’ page, users can access a detailed tutorial about how to use Lnc2Cancer 3.0.

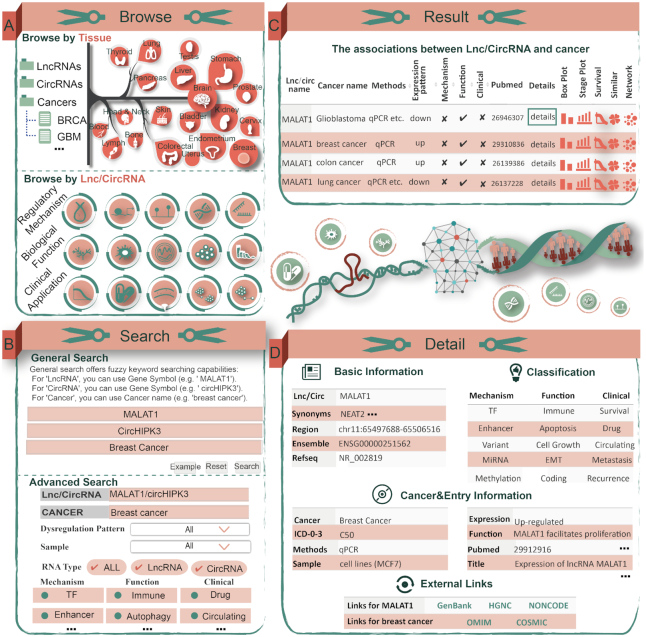
Workflow and case study of basic functions of Lnc2Cancer 3.0. (A) The interface of the browse module, ‘LncRNA-centric’ page and ‘Cancer-centric’ page. (B) The interface of the general search and advanced search modules using MALAT1, circHIPK3 and breast cancer as examples. (C) Query results for MALAT1 in cancer. (D) Basic information, classification, cancer type and entry information for MALAT1 in breast cancer.

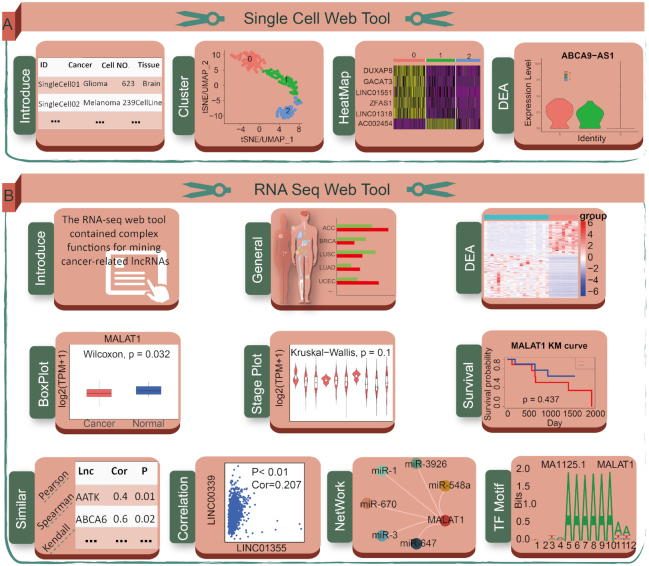
Workflow and case study using web tools in Lnc2Cancer 3.0. (A) Single cell web tools including general information, clustering, heatmap and differential expression analysis for lncRNAs. (B) RNA-seq web tool page including general information, differential expression analysis, box plotting, stage plotting, survival analysis, similar lncRNAs identification, correlation analysis, network construction and TF motif prediction.
CONCLUSIONS AND FUTURE EXTENSIONS
Based on the recent increase in the number of experimentally supported lncRNA-cancer associations (particularly in 2018–2020), we updated and improved the Lnc2Cancer database with the latest data. CircRNAs are a unique type of long non-coding RNAs and are associated with numerous cancers. Thus, we collected experimentally supported cancer-related circRNA entries in Lnc2Cancer 3.0. To the best of our knowledge, the present version of Lnc2Cancer contains the most comprehensive and accurate experimentally supported circRNA-cancer associations. With the advances in the molecular biology research and techniques, we were able to classify and analyze cancer-related lncRNAs and circRNAs based on the regulatory mechanisms, biological functions, and clinical applications. Such classification is valuable for gaining further insights into the roles of lncRNAs and circRNAs in cancer. Notably, the single cell and RNA-seq web tools fill the gap between the available cancer-related lncRNA big data and the delivery of integrated information to end users, thus helping unleash the value of the current data resources concerning the functions of lncRNAs in human cancers. We will continue to maintain and update the Lnc2Cancer database with more datasets and web tools, further improving our understanding of the roles of lncRNAs in cancer.
DATA AVAILABILITY
All the data could be downloaded from http://www.bio-bigdata.net/lnc2cancer/.
FUNDING
National Key R&D Program of China [2018YFC2000100]; National Natural Science Foundation of China [32070672, 32070622, 61873075]; Heilongjiang Touyan Innovation Team Program; Heilongjiang Provincial Natural Science Foundation [LH2020C057]. Funding for open access charge: National Key R&D Program of China [2018YFC2000100].
Conflict of interest statement. None declared.
REFERENCES
2.
3.
5.
6.
7.
8.
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
24.
25.
26.
27.
28.
29.
30.
31.
32.
33.
34.
35.
36.
37.
38.
39.
40.
41.
 Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data
Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data
