
The centralspindlin complex, comprising Tumbleweed RhoGAP and kinesin-like Pavarotti proteins, associates with microtubules during cytokinesis. Nakamura et al. show that Pavarotti has centralspindlin complex–independent functions, binds directly to actin, and regulates actin dynamics during cell wound repair and oogenesis.
Pavarotti, the Drosophila MKLP1 orthologue, is a kinesin-like protein that works with Tumbleweed (MgcRacGAP) as the centralspindlin complex. This complex is essential for cytokinesis, where it helps to organize the contractile actomyosin ring at the equator of dividing cells by activating the RhoGEF Pebble. Actomyosin rings also function as the driving force during cell wound repair. We previously showed that Tumbleweed and Pebble are required for the cell wound repair process. Here, we show that Pavarotti also functions during wound repair and confirm that while Pavarotti, Tumbleweed, and Pebble are all used during this cellular repair, each has a unique localization pattern and knockdown phenotype, demonstrating centralspindlin-independent functions. Surprisingly, we find that the classically microtubule-associated Pavarotti binds directly to actin in vitro and in vivo and has a noncanonical role directly regulating actin dynamics. Finally, we demonstrate that this actin regulation by Pavarotti is not specific to cellular wound repair but is also used in normal development.
Centralspindlin is a conserved heterotetrameric complex composed of dimerized Pavarotti (Pav) and Tumbleweed (Tum) in Drosophila melanogaster (CHO1/MKLP1 and MgcRacGAP in mammals, respectively; D’Avino et al., 2015; Green et al., 2012; Mishima et al., 2002; Pollard and O’Shaughnessy, 2019; Somers and Saint, 2003; White and Glotzer, 2012). Pav is a member of a kinesin-like (kinesin-6) protein family of microtubule (MT)-dependent molecular motors, whereas Tum is a Rho GTPase activating protein that exchanges the GTP-bound form of Rho family GTPase to the GDP-bound form (Adams et al., 1998; Crest et al., 2012; Goldstein et al., 2005; Somers and Saint, 2003; Sommi et al., 2010; Zavortink et al., 2005). During cytokinesis, centralspindlin organizes MTs in the central spindle region, associates with the Pebble (Pbl) Rho guanine nucleotide exchange factor (Ect-2 in mammals), and moves along with Pbl to localize at the equator of the dividing cells (Basant and Glotzer, 2018; D’Avino et al., 2015; Green et al., 2012; Pollard and O’Shaughnessy, 2019; Somers and Saint, 2003; White and Glotzer, 2012). Subsequently, centralspindlin and Pbl activate Rho1, leading to formation of an actomyosin ring at the cortex of the cell equator (Basant and Glotzer, 2018; D’Avino et al., 2015; Green et al., 2012; Pollard and O’Shaughnessy, 2019; Somers and Saint, 2003; White and Glotzer, 2012). Tum, Pav, and Pbl are each essential for the spatial and temporal regulation of Rho1, and their recruitment to the equator is MT dependent (Basant and Glotzer, 2018; Nishimura and Yonemura, 2006; Piekny et al., 2005; Somers and Saint, 2003; Yüce et al., 2005). The role of the centralspindlin complex in cytokinesis has been shown to be highly conserved in a wide variety of model organisms and cell culture models (D’Avino et al., 2015; Green et al., 2012; Pollard and O’Shaughnessy, 2019; White and Glotzer, 2012).
In addition to actomyosin ring formation, the centralspindlin complex regulates MT dynamics during the early stages of cytokinesis, during which it localizes to the antiparallel bundled MTs that compose the spindle midzone (D’Avino et al., 2015; Green et al., 2012; Pollard and O’Shaughnessy, 2019; White and Glotzer, 2012). At later stages, centralspindlin mediates the association of these bundled MTs in the spindle midzone with the plasma membrane to form the midbody structure that is important for completing the final stage of cytokinesis (Lekomtsev et al., 2012). Other examples of MT-dependent or -independent functions of centralspindlin include the migration of neurons in Caenorhabditis elegans, negative regulation of the WNT pathway, and MT sliding, which promotes neurite outgrowth in Drosophila (Del Castillo et al., 2015; Falnikar et al., 2013; Jones et al., 2010). While in most contexts centralspindlin proteins function as this complex, a recent study in C. elegans showed that Rho GTPase activating protein (RhoGAP [Tum]) independently regulates Rho1 activity in oocyte production by the syncytial germline (Lee et al., 2018).
We developed a model to study cell wound repair in the Drosophila syncytial embryo, in which the lateral side of nuclear cycle 4–6 (NC4–6) Drosophila embryos are wounded and the repair process can be visualized in real time by 4D confocal microscopy (Abreu-Blanco et al., 2011a, 2014; Nakamura et al., 2017). This model shares many features of cytokinesis, including an essential actomyosin contractile ring for wound closure and a requirement for MTs (Abreu-Blanco et al., 2011a, 2014; Nakamura et al., 2017, 2018). To date, our work and the work of others has elucidated a cassette of molecular factors involved in cell wound repair that is strikingly similar to those used during cytokinesis (Cooper and McNeil, 2015; Dekraker et al., 2018; Nakamura et al., 2018; Sonnemann and Bement, 2011). In addition to the actomyosin ring, Rho1, Rac1, and Cdc42 are essential to cell wound repair, where they exhibit distinct spatiotemporal patterns (Abreu-Blanco et al., 2014; Benink and Bement, 2005; Nakamura et al., 2017). We recently found that Pbl (Rho guanine nucleotide exchange factor) and Tum (RhoGAP) are recruited to wounds and regulate the spatiotemporal dynamics of actin and myosin, which form the actomyosin ring around the wound edge (Nakamura et al., 2017). Interestingly, however, Pbl and Tum accumulate in distinct spatiotemporal patterns, and Pbl regulates Cdc42 dynamics, rather than those of Rho1, as would be expected from Pbl’s role with the centralspindlin complex in cytokinesis. Additionally, Tum is required for the refinement of Rho1 and Rac1, but not Cdc42, suggesting that Pbl and the centralspindlin complex have separate roles in cell wound repair (Nakamura et al., 2017). Here, we investigate the role of Pav, the other member of the centralspindlin complex, in cell wound repair. We find that, in response to cell wounds, Pav is recruited in a distinct localization pattern, and mutants exhibit a distinct phenotype compared with Tum and Pbl. Importantly, Pav localization at the wound is actin dependent, can directly bind to actin, and functions during wound repair independent from Tum or Pbl. Finally, we demonstrate that Pav regulation of actin dynamics independently from Tum is widespread in Drosophila.
While investigating the signaling pathways upstream of Rho family GTPase localization to cell wounds, we uncovered two lines of evidence suggesting that the centralspindlin complex and its interacting proteins were involved in cell wound repair: (1) Tum and the centralspindlin interacting protein, Pbl, exhibited different localization patterns upon wounding, and (2) Tum and Pbl each had distinct wound repair defects (Nakamura et al., 2017). Because these proteins each had distinct phenotypes and localization patterns, we were interested in determining what role, if any, Tum’s partner Pav plays in cell wound repair to better understand if and how centralspindlin or its constituent proteins were independently involved in cell wound repair.
Upon wounding, actin accumulates in a distinct pattern: (1) a highly enriched actin ring bordering the wound edge and (2) an elevated actin halo encircling the actin ring, which can be used as a reference to orient other protein localizations (Fig. 1 A; Abreu-Blanco et al., 2011a, 2014; Nakamura et al., 2017). As we previously reported, Pbl accumulation only overlaps with the actin halo region, whereas Tum accumulates highly in the actin halo region, and to a lesser extent overlaps with the outer edge of the actin ring (Fig. 1, A–E and K; and Video 1; Nakamura et al., 2017). To assess the localization pattern of Pav, we wounded embryos expressing GFP-tagged Pav driven ubiquitously by the ubiquitin promoter (Minestrini et al., 2002) and expressed at levels similar to its endogenous counterpart, combined with an mCherry-tagged actin reporter (sChMCA; Abreu-Blanco et al., 2011a). Pav accumulates highly at the inner edge of the actin ring and less strongly in the actin halo region (Fig. 1, A, F, G, and J; and Video 1). Interestingly, Pav and Tum accumulation in the actin ring are not identical: Pav most strongly accumulates at the inner actin ring edge, whereas Tum accumulates less strongly and at the outer actin ring edge (Fig. 1, A and D–G; and Video 1). We confirmed this difference by examining embryos coexpressing Ch-tagged Pav and super folder GFP (sfGFP)-tagged Tum (Fig. 1, H and I; Derivery et al., 2015; Nakamura et al., 2017). In addition, we observed similar results when staining wounded embryos expressing sfGFP-Tum with anti-Pav antibodies (anti-Tum antibodies do not work for embryo staining; Fig. 1, L–M””).

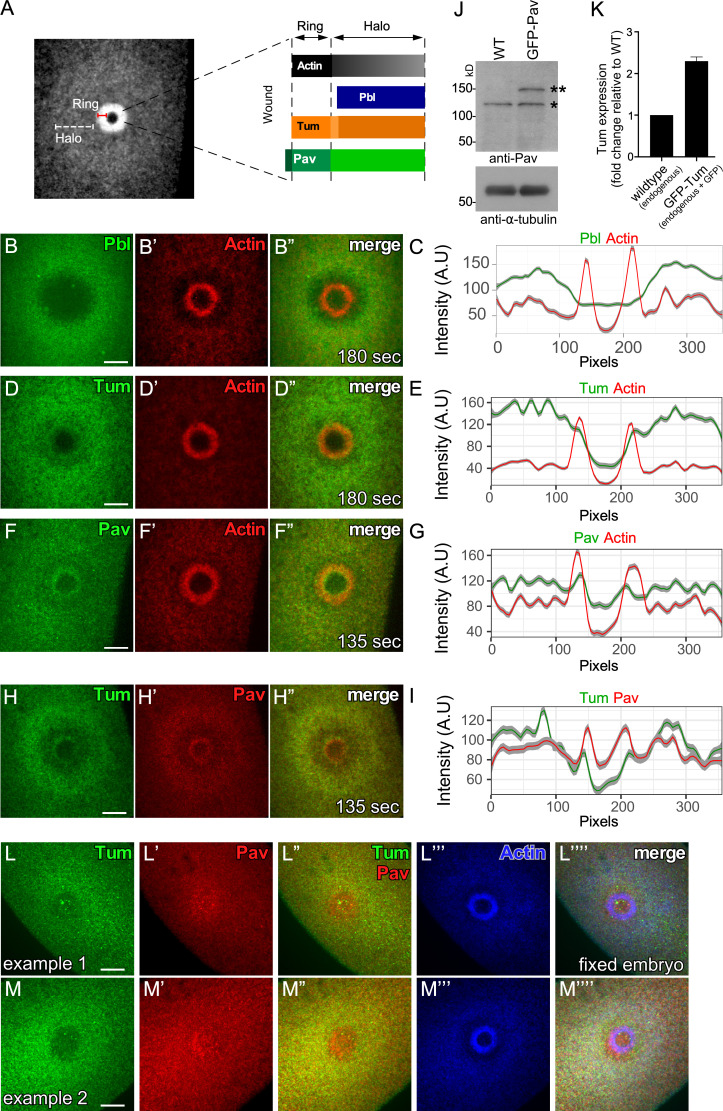
Pbl, Tum, and Pav exhibit distinct localization patterns in cell wound repair. (A) Confocal xy projection image from a laser-wounded NC4–6 staged Drosophila embryo expressing an actin reporter (sGMCA). Schematic diagram summarizing the localization patterns of actin, Pbl, Tum, and Pav at the wound edge. (B–B”) Confocal xy projection images from NC4–6 staged Drosophila embryo coexpressing an actin reporter (sChMCA) and Pbl-eGFP. (C) Fluorescence intensity (arbitrary units) profiles across the wound area in B”. (D–D”) Confocal xy projection images from a NC4–6 staged Drosophila embryo coexpressing an actin reporter (sChMCA) and sfGFP-Tum. (E) Fluorescence intensity (arbitrary units) profiles across the wound area in D”. (F–F”) Confocal xy projection images from an NC4–6 staged Drosophila embryo coexpressing an actin marker (sChMCA) and GFP-Pav. (G) Fluorescence intensity (arbitrary units) profiles across the wound area in F”. (H–H”) Confocal xy projection images from a NC4–6 staged Drosophila embryo coexpressing sfGFP-Tum and Ch-Pav. (I) Fluorescence intensity (arbitrary units) profiles across the wound area in H”. (J) Western blot analysis of Pav protein levels in wild type and GFP-Pav–expressing embryos under our imaging conditions (*, endogenous Pav; **, GFP-Pav). (K) qPCR analysis of Tum expression in wild-type and GFP-Tum expressing embryos under our imaging conditions. Error bars represent ± SEM. (L–M””) GFP-Tum–expressing embryos stained for Tum (anti-GFP), Pav (anti-Pav), and F-actin (phalloidin). Scale bar: 20 µm. Time after wounding is indicated. In fluorescence intensity profiles, the line represents the averaged fluorescent intensity, and the gray area is the 95% confidence interval from a 10-pixel section of the image shown (B”, D”, F”, or H”).
Pav and Tum exhibit distinct localization patterns in cell wound repair. (A and B) Time-lapse confocal xy images and fluorescence intensity (arbitrary units) profiles across the wound area from Drosophila NC4–6 staged embryos coexpressing an actin reporter (sChMCA, red) along with sfGFP-Tum (green; A) or GFP-Pav (green; B). (C) Time-lapse confocal xy images from Drosophila NC4–6 staged embryos coexpressing sfGFP-Tum (green) and Ch-Pav (red). Time after wounding is indicated. Playback rate is 10 frames/s.
We next compared the wound repair phenotypes in tum and pav knockdown backgrounds using a fluorescent actin reporter (sChMCA or GFP-tagged actin reporter [sGMCA]; Kiehart et al., 2000) that, in the knockdown background, allows us to assay physical properties of the repair process (i.e., size, expansion, and closure rate) and defects in actin structures during this process. We generated pav knockdown embryos two different ways: (1) expressing two independent RNAi constructs for pav (HMJ02232 and GL01316) in the female germline using the GAL4-UAS system, and (2) using the wimp mutation to generate reduced Pav expression in both the germline and soma (maternal contribution of Pav is reduced in trans-heterozygotes of pav and wimp, referred as reduced pav; Parkhurst and Ish-Horowicz, 1991). Unfortunately, females with all combinations of maternal GAL4s and RNAi lines do not produce any eggs. However, reduced pav mutants from two independent alleles (pav963 and pavB200) show significantly decreased protein levels (≥95% knockdown; Fig. 2 A) and produce embryos that can be used to discern Pav function in wound repair. While the spatiotemporal regulation of Rho family GTPases is indispensable for actomyosin ring formation during cell wound repair and the centralspindlin complex is required to activate Rho1 during cytokinesis, Pav is not required for recruitment of Rho family GTPases to wounds during cell wound repair (Fig. S1 and Video 2). Both reduced pav963 and reduced pavB200 embryos exhibit similar wound repair phenotypes: a narrow actin ring, delayed actin accumulation around the wound edge, and a slower closure rate compared with control (wimp/+) embryos (Fig. 2, C–E’, J, K, and N–P; and Video 3). In contrast to reduced pav mutants, tum knockdown embryos obtained from RNAi knockdown with either of two independent lines, HMS05764 (tumRNAi(1); Fig. 2 B) and UAS-RacGAP50C-dsRNA (tumRNAi(2); Nakamura et al., 2017) exhibit a different phenotype: a wider actin ring, wound overexpansion, and a slightly faster closure rate (Fig. 2, F–I, L–N, P, and Q; and Video 3). In dividing cells, Tum depletion causes Pav to no longer be recruited to the equator (Zavortink et al., 2005; Tao et al., 2016). Surprisingly, we found that Pav and Tum are still recruited to wounds in the absence of the other (Fig. 2, R–U). Taken together, our localization and mutant analyses suggest centralspindlin complex–independent roles for Pav and Tum during wound repair.

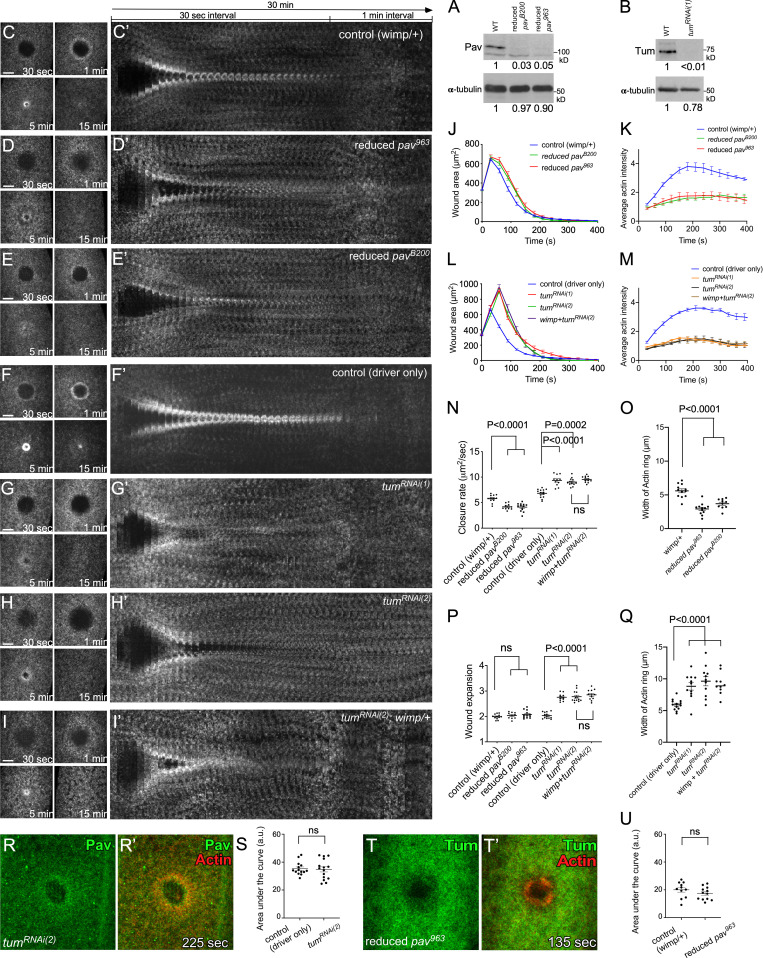
Pav and Tum mutants exhibit distinct phenotypes. (A) Western blot analysis of Pav protein levels (100 kD) in lysates from wild type (WT), reduced pavB200, and reduced pav963. (B) Western blot analysis of Tum protein levels (70 kD) in lysates from WT and tumRNAi(1). (C–I’) Actin dynamics (sGMCA or sChMCA) during cell wound repair in NC4–6 staged embryos: control (wimp/+; C), reduced pav963 (wimp+/+ pav963; D), reduced pavB200 (wimp+/+ pavB200; E), control (GAL4 driver alone; F), tumRNAi(1) (G), tumRNAi(2) (H), tumRNAi(2) : wimp/+ (I). (C’–I’) xy kymograph across the wound area depicted in C–I, respectively. (J) Quantification of the wound area over time for control (wimp/+; n = 12), reduced pav963 (n = 12), and reduced pavB200 (n = 10). (K) Quantification of the average intensity within the actin ring over time relative to UW for control (wimp/+; n = 5), reduced pav963 (n = 5), and reduced pavB200 (n = 5). (L) Quantification of the wound area over time for control (GAL4 driver alone; n = 14), tumRNAi(1) (n = 11), tumRNAi(2) (n = 13), and tumRNAi(2) : wimp/+ (n = 12). (M) Quantification of the average intensity within the actin ring over time relative to UW for control (GAL4 driver alone; n = 5), tumRNAi(1) (n = 5), tumRNAi(2) (n = 5), and tumRNAi(2) : wimp/+ (n = 5). (N–Q) Quantification of wound closure speed (N), wound expansion time (P), and actin ring width (O and Q) for control (wimp/+), reduced pav963, reduced pavB200, control (GAL4 driver alone), tumRNAi(1), tumRNAi(2), and tumRNAi(2) : wimp/+. (R–R’) Confocal xy projection images from a NC4–6 staged Drosophila embryo coexpressing an actin marker (sChMCA) and GFP-Pav in a tumRNAi(2) background. (S) Quantification of the area under the curve in each fluorescence intensity profile form control (GAL4 driver only; n = 14) and tumRNAi(2) (n = 15). ns, not significant. (T–T’) Confocal xy projection images from a NC4–6 staged Drosophila embryo coexpressing an actin marker (sStMCA) and sfGFP-Tum in a reduced pav963 background. (U) Quantification of the area under the curve in each fluorescence intensity profile form control (wimp/+; n = 11) and reduced pav963 (n = 12). Time after wounding is indicated. Scale bar: 20 µm. Error bars represent ±SEM. Unpaired Student’s t tests were performed in N–Q, S, and U).

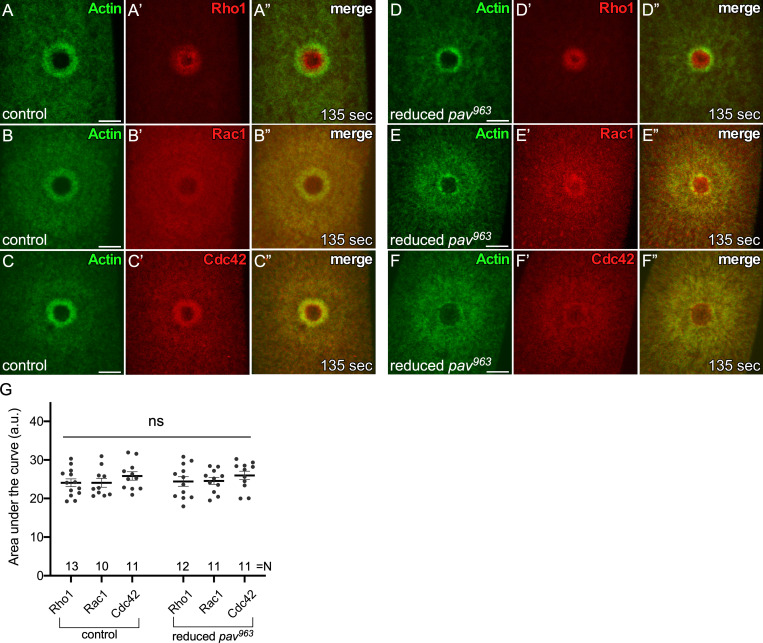
Spatial and temporal patterns of Rho family GTPases are not altered in reduced pav mutant backgrounds. (A–C”) Localization of Rho family GTPases along with an actin reporter (sGMCA) in NC4–6 staged control embryos (wimp/+): ChFP-Rho1 (A–A”), ChFP-Rac1 (B–B”), and ChFP-Cdc42 (C–C”). (D–F”) Localization of Rho family GTPases along with an actin reporter (sGMCA) in NC4–6 staged reduced pav embryos: ChFP-Rho1 (D–D”), ChFP-Rac1 (E–E”), and ChFP-Cdc42 (F–F”). (G) Quantification of the area under the curve in each fluorescence intensity profile from 10–12 individual embryos. ns, not significant. Unpaired Student’s t tests were performed in G. n and time after wounding are indicated. Scale bar: 20 µm. Error bars represent ±SEM.
Rho family GTPases are recruited to cell wounds in reduced pav mutants. (A–F) Time-lapse confocal xy images from Drosophila NC4–6 staged embryos coexpressing an actin reporter (sGMCA, green) along with Rho family GTPase in control (wimp/+; A–C) and reduced pav (D–F): Ch-Rho1 (A and D, red), Ch-Rac1 (B and E, red), and Ch-Cdc42 (C and F, red). Time after wounding is indicated. Playback rate is 10 frames/s.
Pav and Tum mutants exhibit distinct phenotypes in cell wound repair. (A–G) Time-lapse confocal xy images from Drosophila NC4–6 staged embryos expressing an actin marker (sGMCA or sChMCA): control (wimp/+; A), reduced pavB200 (B), reduced pav963 (C), control (Gal4 driver/+; D), tumRNAi(1) (E), tumRNAi(2) (F), and tumRNAi(2)+wimp (G). (H and I) Time-lapse confocal xy images and fluorescence intensity (arbitrary units) profiles across the wound area from Drosophila NC4–6 staged embryos coexpressing an actin reporter (sChMCA [H] or sStMCA [I], red) along with GFP-Pav (H) or sfGFP-Tum (I) in tumRNAi(2) background (H) or reduced pav963 (I). Time after wounding is indicated. Playback rate is 10 frames/s.
Kinesin-like proteins associate with MTs and transport cargo proteins (Hirokawa et al., 2009; Lu and Gelfand, 2017; Vale, 2003); thus, we expected that disrupting MTs would affect Pav recruitment to wounds. We previously found that although we did not observe an accumulation or rearrangement of MTs around wounds, the presence of the MT network is required for proper wound repair, as injection of colchicine (inhibitor of MT polymerization) into embryos disrupted actin dynamics (Abreu-Blanco et al., 2011a). Using a new MT reporter (Maple3-α-tubulin; Lu et al., 2016) and improved microscopic technologies, we found that MTs are uniformly distributed at wounds from the internal membrane plug region through to the outer actin halo region, with decreased uniformity at the outermost edge of the actin halo (Fig. 3, A–A”; and Video 4). This MT distribution at wounds is different from the accumulation of Pav and is disrupted upon injecting colchicine, latrunculin B (LatB; inhibitor of actin polymerization), and colchicine + LatB (Fig. 3, B–D”; and Video 4), indicating that actin and MT cytoskeletons are intimately intertwined during cell wound repair. To examine whether the function of Pav is required for actin dynamics, we wounded embryos coexpressing an actin reporter (sChMCA) and Pav-GFP upon injecting buffer, colchicine, and LatB. Although Pav accumulation at the edge of the actin ring is disrupted in colchicine-injected embryos compared with buffer control, Pav is still recruited to wounds, and its accumulation overlaps that of actin (Fig. 3, E–J”; and Video 5). Surprisingly, Pav localization is severely disrupted in LatB-injected embryos and largely mirrors the disrupted actin structures (Fig. 3, G–H”; and Video 5). Because some GFP-Pav puncta do not overlap with actin, we also examined Pav localization upon injecting colchicine and LatB simultaneously, and in this case found that Pav still overlaps with the disrupted actin structures (Fig. 3, I–J”; and Video 5). In addition to Pav, we examined Tum localization upon wounding in the presence or absence of the same cytoskeleton inhibitors. We found that Tum is also recruited to wounds upon colchicine injection but is severely disrupted by LatB injection (Fig. S2, A–D”; and Video 4). Our results indicate that Pav and Tum localization is dependent on actin, rather than MTs, in the context of cell wound repair.

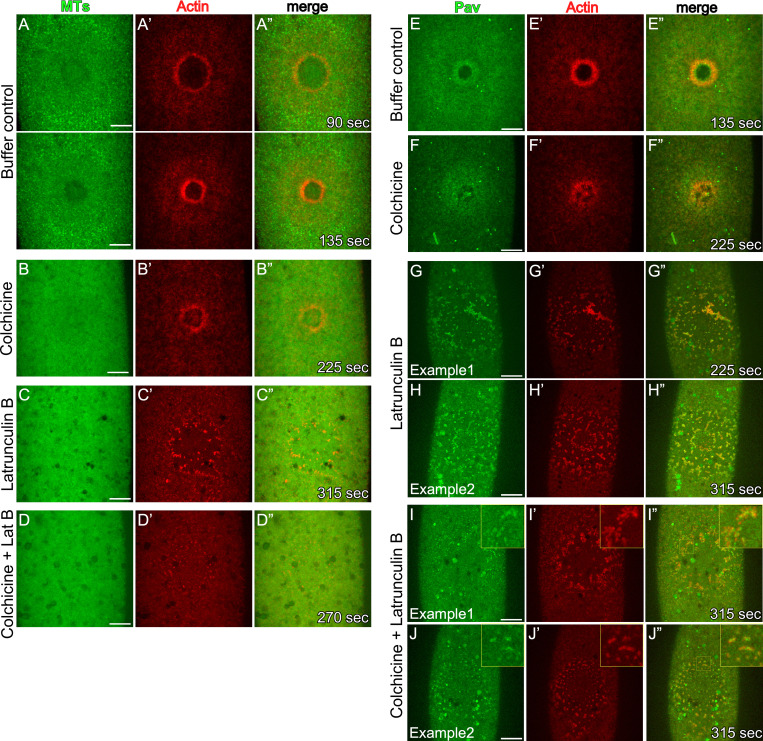
GFP-Pav overlaps with actin during cell wound repair. (A–D”) Localization of α-tubulin and an actin reporter (sStMCA) upon injecting buffer control (A–A”), colchicine (B–B”), LatB (C–C”), or colchicine + LatB (D–D”) in NC4–6 staged embryos. (E–J”) Localization of GFP-Pav and an actin reporter (sChMCA) upon injecting buffer control (E–E”), colchicine (F–F”), LatB (G–H”), or colchicine + LatB (I–J”) in NC4–6 staged embryos. Time after wounding is indicated. Scale bar: 20 µm.
MT and Tum dynamics during cell wound repair following drug injection. (A–H) Time-lapse confocal xy images from Drosophila NC4–6 staged embryos coexpressing an actin reporter (sStMCA, red) with α-tubulin-Maple3 (green; A–D) or sfGFP-Tum (green; E–H) upon injecting buffer (A and E), colchicine (B and F), LatB (C and G), or colchicine + LatB (D and H). Time after wounding is indicated. Playback rate is 10 frames/s.
GFP-Pav overlaps with actin during cell wound repair. (A–F) Time-lapse confocal xy images from Drosophila NC4–6 staged embryos coexpressing an actin reporter (sChMCA, red) with GFP-Pav (green) upon injecting buffer (A), colchicine (B), LatB (C and D), or colchicine + LatB (E and F). Time after wounding is indicated. Playback rate is 10 frames/s.

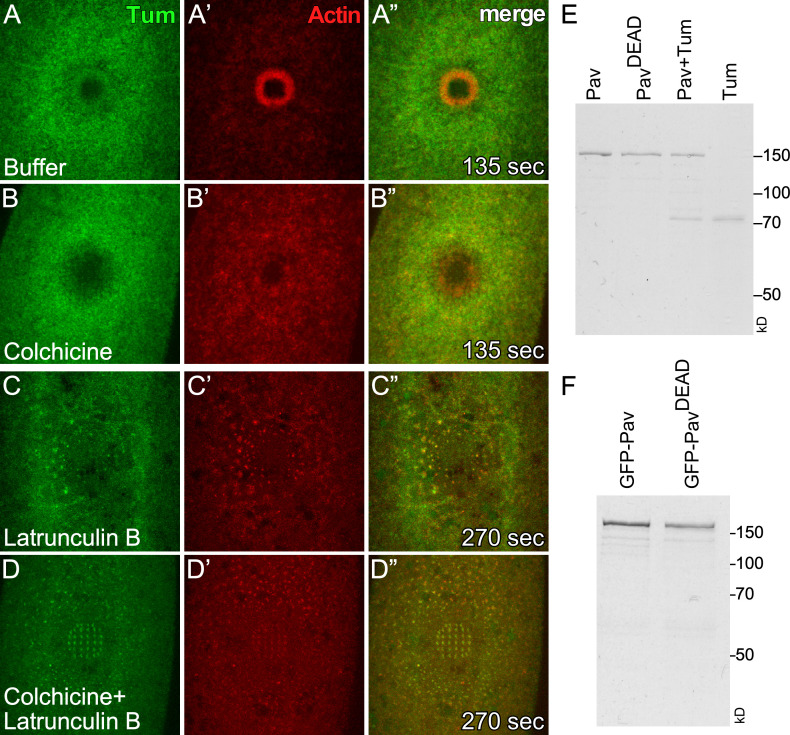
Tum recruitment to wounds depends on actin. (A–D”) Localization of sfGFP-Tum and an actin reporter (sChMCA) upon injecting buffer control (A–A”), colchicine (B–B”), LatB (C–C”), or colchicine + LatB (D–D”) in NC4–6 staged embryos. Time after wounding is indicated. Scale bar: 20 µm. (E) Coomassie-stained SDS-PAGE gels of proteins used in this study.
Consistent with Pav’s actin-dependent localization, a previous study identified an actin binding site in the 18th exon of CHO1, one of two isoforms of MKLP1, the mammalian homologue of Pav (Kuriyama et al., 2002). Although alignment reveals that this actin binding site is not conserved in Pav, it is possible that Drosophila Pav still encodes actin binding activity. To determine whether Pav binds directly to F-actin, we first performed low-speed cosedimentation of bacterially purified full-length Pav protein. Unfortunately, Pav protein pellets without F-actin and/or MTs present, perhaps owing to its large size (>100 kD; Fig. S2 E) and/or dimerization (not depicted; Basant et al., 2015; Hutterer et al., 2009; Zhou et al., 2010). Hence, we performed in vitro F-actin and MT bundling/cross-linking assays (Fig. 4 A). In vitro polymerized F-actin or MTs distribute uniformly without organization and are not bundled or cross-linked in the absence of additional factors (Fig. 4, B and B’). Addition of bacterially purified full-length Pav protein bundles MTs, as reported previously (Tao et al., 2016), in the presence or absence of F-actin (Fig. 4, C’, C”, and E; and Fig. S3, B–E’). Surprisingly, addition of Pav protein also bundles F-actin, both alone and in the presence of MTs (Fig. 4, C and D; and Fig. S3, B–E). In addition to Pav bundling actin and MTs, these bundles are linked in that they overlap with each other, suggesting that Pav mediates actin/MT cross-linking (Fig. 4, C–C”; and Fig. S3, B–E”).

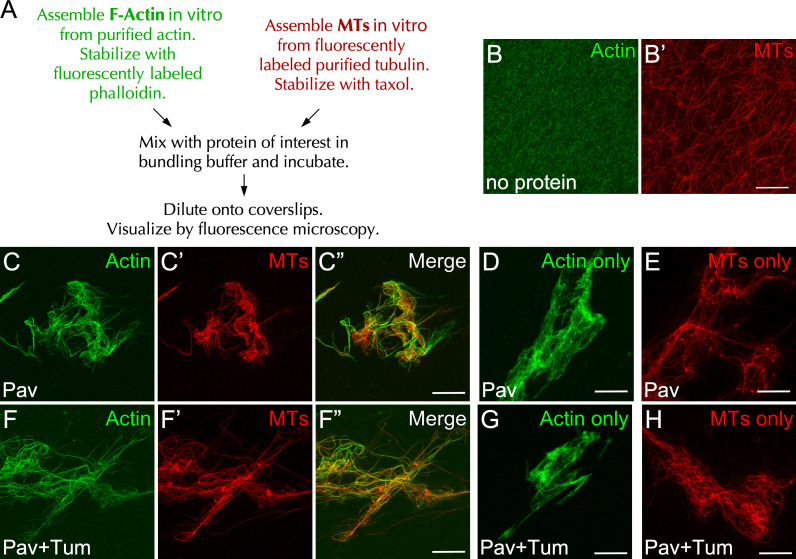
Pav bundles F-actin/MTs and cross-links them, even in the presence of Tum. (A) The scheme of bundling/cross-linking assays. (B–H) Stabilized actin and/or MTs were incubated with no protein (B and B’), full-length Pav protein, or full-length Pav/Tum proteins (C–H). Final protein concentrations for bundling assays: Pav, 200 nM; Pav + Tum, 200 nM Pav. Scale bar: 30 µm.

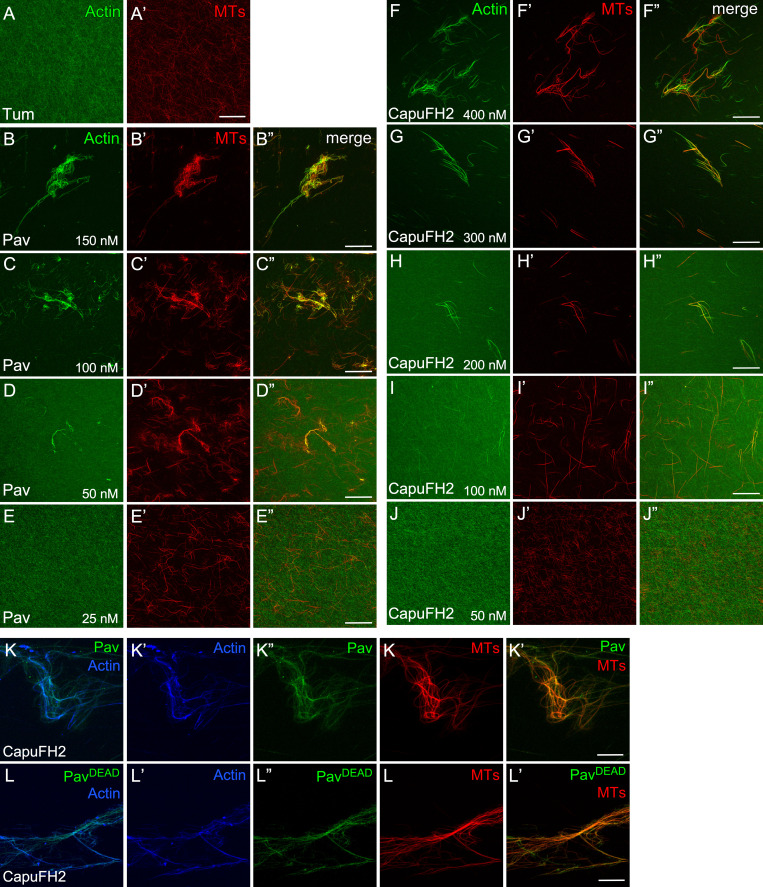
Pav and CapuFH2 bundle and cross-link actin/MTs in a concentration-dependent manner. (A and A’) Stabilized actin and MTs incubated with Tum. (B–J”) Stabilized actin and/or MTs were incubated with different concentrations of Pav (B–E”) and CapuFH2 (F–J”). Protein concentration is indicated. (K–L””) Stabilized actin and MTs were incubated with CapuFH2 to bundle actin/MTs, followed by the addition of GFP-Pav (K–K””) or GFP-PavDEAD (L–L’”). Scale bar: 30 µm. Final protein concentrations for bundling assays: CapuFH2, 500 nM; GFP-Pav, 3 nM; and GFP-PavDEAD, 3 nM.
The aforementioned study in Drosophila also showed that Tum works as a molecular toggle to activate the kinesin motor function of Pav (Tao et al., 2016), and because we observed that Tum overlaps with actin during cell wound repair, we next examined whether the association of Tum to Pav could affect the F-actin/MT bundling and/or cross-linking activities of Pav. Copurified Pav and Tum proteins from bacterial lysates (see Materials and methods) did not affect the bundling and/or cross-linking activities of Pav (Fig. 4, F–H). In addition, purified Tum alone did not bundle and/or cross-link actin/MTs (Fig. S3, A and A’). Thus, Pav can bind directly to actin/MTs and cross-link them regardless of the presence of Tum and without additional factors.
Pav has three major domains: the head, stalk, and tail domains (Fig. 5 A; Minestrini et al., 2002). The MT binding activity of MKLP1/CHO1 is located in its head domain (Kuriyama et al., 2002; Matuliene and Kuriyama, 2002). To map the region of Drosophila Pav required for its actin-binding activity, we purified four Pav protein fragments containing the head domain, the head + stalk domains, the stalk + tail domains, and the tail domain (Fig. 5 A), and then assayed their activities in bundling assays. Interestingly, the Pav head domain bundles actin but not MTs, whereas the Pav head + stalk domains can bundle/cross-link both actin and MTs (Fig. 5, B and C”). The Pav stalk + tail domains and Pav tail domain do not bundle either of them (Fig. 5, D–E’). This suggested that the combination head + stalk domains is necessary for actin/MT bundling/cross-linking.

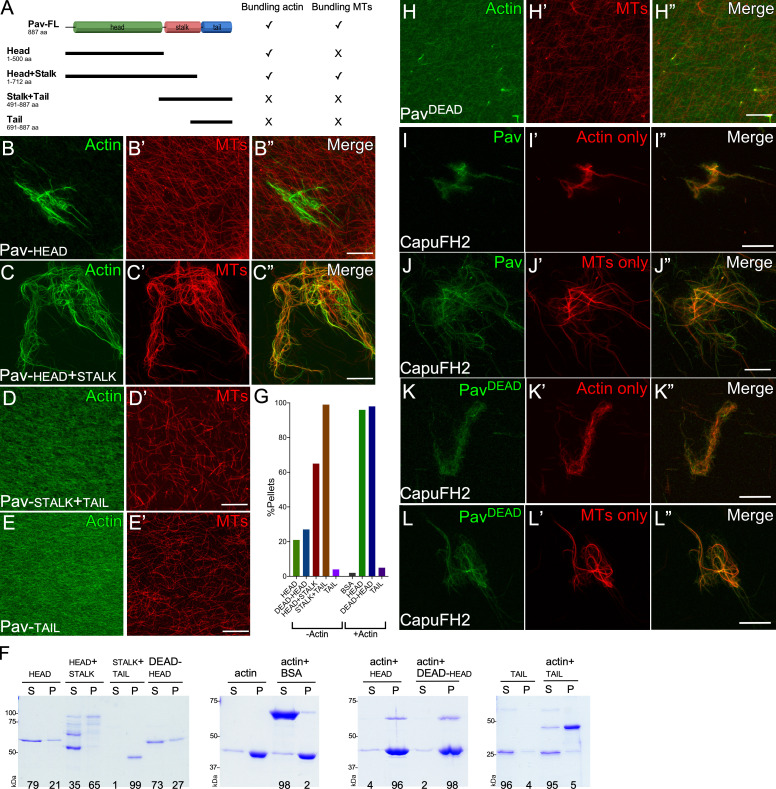
Pav binds to actin through its head region. (A) Schematic diagrams illustrating the protein domains of Pav and different pieces of Pav used in bundling and cosedimentation assays. (B–E”) Stabilized actin and/or MTs were bundled and cross-linked by Pav-head (B–B”), Pav-head + stalk (C–C”), Pav-stalk + tail (D and D’), and Pav-tail (E and E’). (F) High-speed cosedimentation assay of actin in the presence of different Pav protein pieces. The percentage of Pav protein segregating with the pellets or remaining in the supernatant is shown at the bottom of gel. (G) Quantification of Pav proteins pelleting in the presence or absence of actin. (H–H”) Stabilized actin and/or MTs were incubated with PavDEAD. (I–L”) Stabilized actin or MTs were incubated with CapuFH2 to bundle actin or MTs, followed by the addition of GFP-Pav (I–J”) or GFP-PavDEAD (K–L”). Final protein concentrations for bundling assays: Pav-head, 1 µM; Pav-stalk + tail, 1 µM; Pav-head + stalk, 300 nM; Pav-tail, 2 µM; Pav-PavDEAD, 200 nM; CapuFH2, 500 nM; GFP-Pav, 3 nM; and GFP-PavDEAD, 3 nM. Final protein concentrations for cosedimentation assays: Pav-head, 300 nM; Pav-stalk + tail, 300 nM; Pav-head + stalk, 200 nM; Pav-tail, 800 nM; PavDEAD-head, 300 nM; BSA, 2 µM; and actin, 2 µM. Scale bar: 30 µm.
To quantify the actin binding activity of these partial Pav proteins, we performed high-speed actin cosedimentation assays (Fig. 5, F and G). Because the head + stalk and the stalk + tail proteins pellet without F-actin, we used the Pav head and tail domain for high-speed actin cosedimentation assays. The Pav head domain pellets in the presence of F-actin, whereas the Pav tail domain and BSA (a negative control) do not (Fig. 5, F and G), indicating that the Pav head domain contains the actin binding site.
It has previously been shown that MT binding by the head domain of kinesins depends on nucleotide, and mutation of the kinesin ATP binding site alters the protein to a rigor-like state (Kull and Endow, 2013). Specifically, a G131E mutation in Pav’s head domain (referred to as PavDEAD) exhibits rigor-like association to and stabilization of MTs in vivo (Minestrini et al., 2002, 2003). To examine whether this Pav G131E mutation could affect Pav’s actin binding/bundling activity, we purified PavDEAD protein. PavDEAD protein has decreased capacity for bundling actin and MTs compared with its wild-type counterpart (Fig. 5, B–B” and H–H”).
We next determined whether the PavDEAD protein loses its ability to bundle actin because of a decreased ability to bind actin. We added GFP-tagged Pav or PavDEAD protein to bundled/cross-linked actin and/or MTs. Importantly, the formin Cappuccino was used to bundle/cross-link actin and MTs so that the system was Pav independent before addition of the GFP-tagged Pav proteins (Fig. S3, F–J”; Rosales-Nieves et al., 2006). Localization of GFP-tagged PavDEAD protein on actin/MTs, actin alone, and MTs alone (Fig. 5, K–L”; and Fig. S3, K–K””) is similar to GFP-tagged Pav (Fig. 5, I–J”; and Fig. S3, L–L””), suggesting that the PavDEAD protein still can bind to actin and MTs. Consistent with this, purified PavDEAD-head protein has similar binding activity compared with Pav head protein (for n = 5: Pav-head pellets, 84 ± 7%; PavDEAD-head pellets, 87 ± 5%) in high-speed actin cosedimentation assays (Fig. 5, F and G). Taken together, our results show that PavDEAD protein loses actin/MT bundling activity but retains its ability to bind to actin.
We were interested in determining whether this newly appreciated actin-binding capacity of Pav was important for processes other than cell wound repair. Previous studies had indicated that Pav and Tum are colocalized at ring canals during oogenesis (Airoldi et al., 2011; Mannix et al., 2019; Minestrini et al., 2002). Ring canals, comprising inner and outer actin-rich rings, are created by incomplete cytokinesis during Drosophila oogenesis and function to connect the oocyte and nurse cells (Hudson and Cooley, 2002; Ong and Tan, 2010; Robinson et al., 1994; Warn et al., 1985; Fig. 6, A–C’”). Here, Pav and Tum work together as the centralspindlin complex. However, maternal knockdown of Tum results in embryos, whereas maternal knockdown of Pav does not, suggesting that Pav might have additional centralspindlin-independent functions during oogenesis. To determine whether Pav functions solely as part of the centralspindlin complex during oogenesis, we examined Tum and Pav localization in stage 7 egg chambers using flies expressing sfGFP-tagged Tum (driven by the ubiquitous spaghetti squash promoter) and staining for Pav and GFP (Nakamura et al., 2017). We found that while Tum and Pav are both present throughout the oocyte and nurse cells, they showed specific enriched localization at inner ring canals and in nurse cell nuclei (Fig. 6, A–C’”). Interestingly, Pav also shows specific enrichment with the outer actin ring, and this Pav enrichment is lost in Pav-knockdown backgrounds (Fig. 6, B–C’”; and Fig. S4, A–B”). We also observed Pav, but not Tum, accumulation at the actin-rich oocyte cortex (Fig. 6, D–F”). Accumulation of both Tum and Pav at the ring canals is not surprising, as these were once sites of cytokinesis; however, we were surprised that their localization was only partially overlapping.

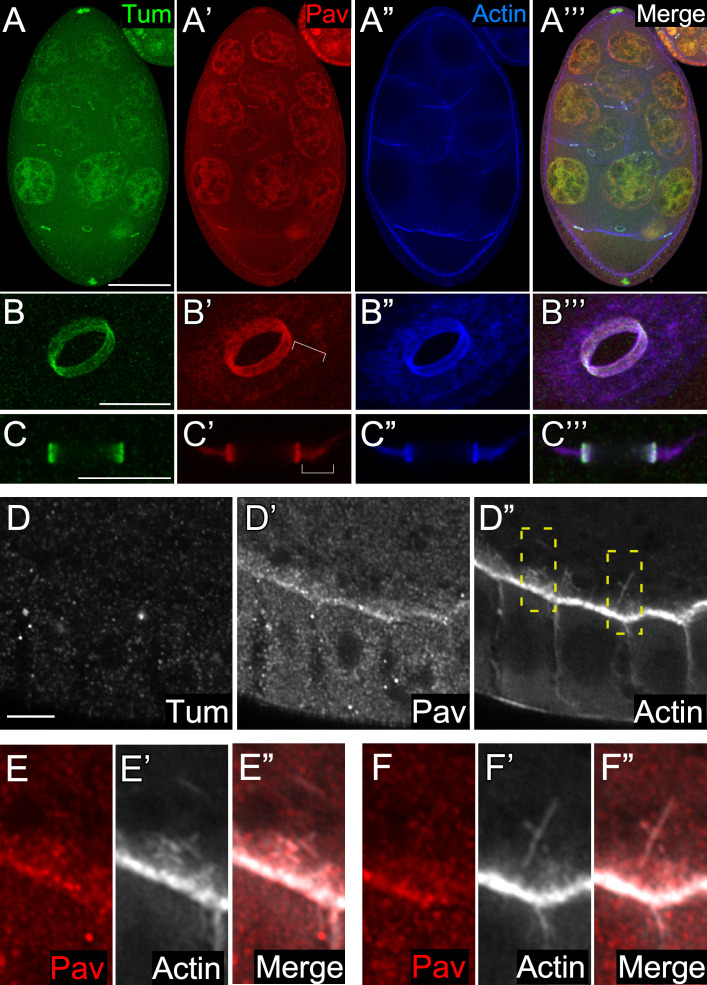
Pav and Tum also exhibit independent localizations in the ovary. (A–A’”) GFP-Tum–expressing stage 7 egg chamber stained for Tum (anti-GFP), Pav (anti-Pav), and F-actin (phalloidin). Scale bar: 50 µm. (B–C’”) Oblique view of the ring canal connecting two nurse cells (B–B’”) and cross section of the ring canal connecting an oocyte and a nurse cell (C–C’”) in the stage 7 egg chambers. Brackets indicate outer actin ring canals. Scale bar: 10 µm. (D–D”) Protein distribution at the GFP-Tum–expressing oocyte cortex in the stage 7 egg chamber, stained for Tum (anti-GFP), Pav (anti-Pav), and F-actin (phalloidin). Scale bar: 10 µm. (E–F”) High-magnification views from two different boxed areas in D”. Scale bar: 1 µm.

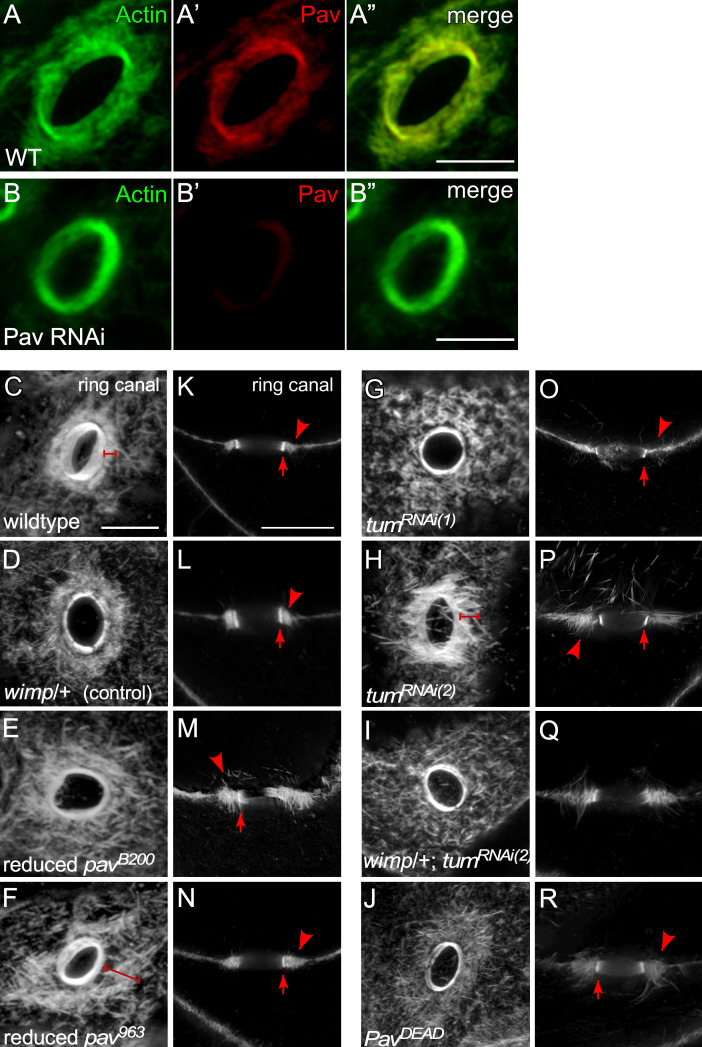
Pav and Tum are required for the organization of actin in outer ring canals. (A–B”) Oblique view of the ring canal connecting two nurse cells in the stage 7 egg chambers, stained for anti-Pav and F-actin in wild type (A–A”) and Pav RNAi mutant (B–B”). (C–J) Oblique view of the ring canal connecting two nurse cells in stage 7 egg chambers, stained for F-actin, in wild type (C), wimp/+ (D), reduced pavB200 (E), reduced pav963 (F), tumRNAi(1) (G), tumRNAi(2) (H), and tumRNAi(2) : wimp/+ (I): PavDEAD (J). Brackets indicate outer ring canal. (K–R) Cross section of the ring canal connecting an oocyte and a nurse cell in stage 7 egg chamber, stained for F-actin, in wild type (K), wimp/+ (L), reduced pavB200 (M), reduced pav963 (N), tumRNAi(1) (O), tumRNAi(2) (P), and tumRNAi(2) : wimp/+ (Q): PavDEAD (R). Arrow indicates inner ring canal; arrowhead indicates outer ring canal. Scale bars: 10 μm.
Actin and MTs have distinct functions during oogenesis: MTs are important for polarity/axis formation by transporting mRNA/proteins and for ooplasmic streaming, whereas actin is required for the formation of ring canals and the timing of ooplasmic streaming (Bastock and St Johnston, 2008; Haglund et al., 2011; McLaughlin and Bratu, 2015; Quinlan, 2016; Robinson and Cooley, 1996). Hence, we examined ring canal morphology in reduced pav mutant and tum RNAi ovaries. Based on their partially nonoverlapping localizations, we suspected that Tum and Pav have centralspindlin complex–independent functions and would display both common and unique phenotypes. Reduced pav mutants have a slightly disorganized inner actin ring with a broad, detached, and highly disorganized outer ring canal with thick actin filaments protruding into the cells (Fig. 7 A and Fig. S4, C–R). In contrast to this, tum RNAi knockdowns show a mildly disorganized inner actin ring and prominent actin filaments into the nurse cells, and the F-actin filaments of the outer ring canal bind to the inner ring canal perpendicular to their normal orientation (Fig. 7 A and Fig. S4, C–R). Both protein localization and mutant phenotypes of Tum and Pav suggest that these proteins are acting at least partially independently in this context.

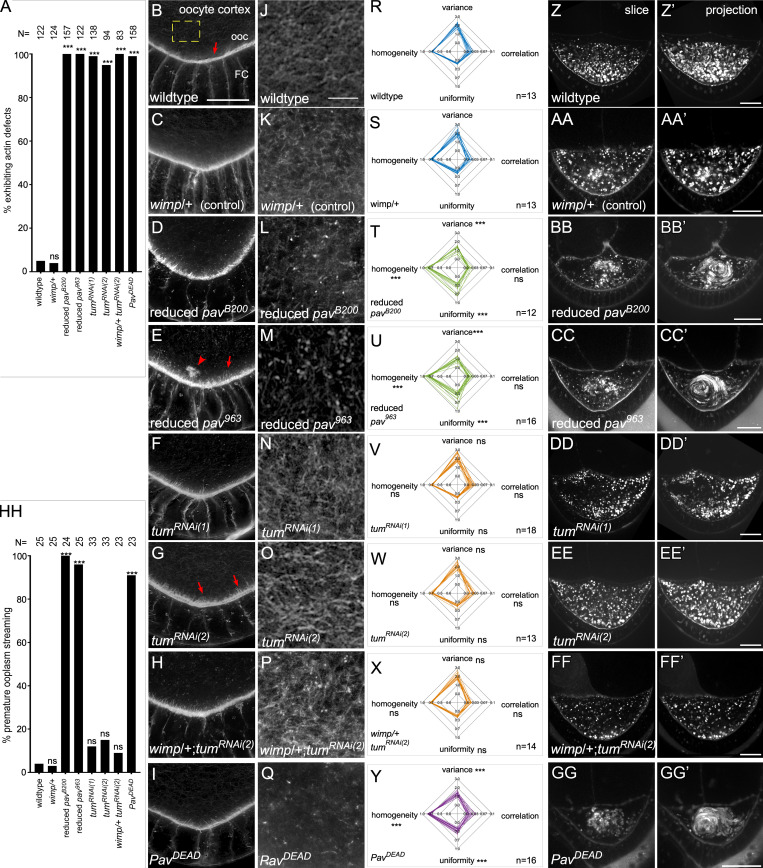
Actin-dependent Pav function is required for oogenesis. (A) Quantification of the percentage of egg chambers exhibiting actin defects. (B–I) Posterior of stage 7 egg chambers, stained for F-actin, in wild type (B), wimp/+ (C), reduced pavB200 (D), reduced pav963 (E), tumRNAi(1) (F), tumRNAi(2) (G), and tumRNAi(2) : wimp/+ (H): PavDEAD (I). Arrows indicate cortical actin projections from oocyte (ooc) cortex. Arrowhead indicates actin aggregation in the oocyte. (J–Q) Cortical actin mesh structure in wild type (J), wimp/+ (K), reduced pavB200 (L), reduced pav963 (M), tumRNAi(1) (N), tumRNAi(2) (O), and tumRNAi(2) : wimp/+ (P): PavDEAD (Q). (R–Y) Quantification of actin mesh structure with GLCM texture analysis for wild type (R), wimp/+ (S), reduced pavB200 (T), reduced pav963 (U), tumRNAi(1) (V), tumRNAi(2) (W), and tumRNAi(2) : wimp/+ (X): PavDEAD (Y). (Z–GG’) Single time point (Z–GG) and 30-min time point projections (Z’–GG’) from time-lapse movies of stage 7 egg oocytes in wild type (Z and Z’), wimp/+ (AA and AA’), reduced pavB200 (BB and BB’), reduced pav963 (CC and CC’), tumRNAi(1) (DD and DD’), tumRNAi(2) (EE and EE’), tumRNAi(2) : wimp/+ (FF and FF’), and PavDEAD (GG and GG’). (HH) Quantification of the percentage of egg chambers exhibiting premature ooplasmic streaming. n for each genotype is indicated on bar plots; Fisher’s exact test was performed in A and HH, and unpaired Student’s t test was performed in R–Y; ns, not significant. ***, P < 0.0001. Scale bars: 20 µm in Z–GG’ and 10 µm in B–Q.
We and others have previously shown that actin/MT bundling and cross-linking at the oocyte cortex, as well as a cytoplasmic actin mesh, are essential to prevent premature ooplasmic swirling (Dahlgaard et al., 2007; Liu et al., 2009; Lu et al., 2016; Rosales-Nieves et al., 2006; Wang and Riechmann, 2008). As Pav binds F-actin filaments in vitro (Fig. 5, C and D), we used high-resolution microscopy to determine that Pav also localizes to F-actin structures in vivo at the oocyte cortex (Fig. 6, D–F”). Consistent with this, Pav affects actin organization during oogenesis (Fig. 7 A). In control oocytes, cortical actin is highly organized, with one layer of uniformly sized spike-like structures (Fig. 7, B and C). Reduced pav mutants exhibit disrupted cortical actin organization, resulting in abnormally long actin filaments and actin aggregations within the ooplasm (Fig. 7, D and E). Intriguingly, tum RNAi oocytes exhibited abnormal cortical actin, albeit less severely than reduced pav, with long actin filaments and separated cortex layers, despite no evidence of specific accumulation at this region (Fig. 7, F–H). This subtle phenotype may be indirect and attributable to the loss of Tum in other parts of the egg chamber. To further examine the different effects of Pav and Tum on actin regulation, we quantified their effects on the actin mesh in stage 8 oocytes using gray level co-occurrence matrix (GLCM) texture analysis with four parameters: variance/contrast, correlation, uniformity, and homogeneity (Fig. 7, J–Y; see Materials and methods; Haralick et al., 1973). The actin mesh in tum RNAi oocytes was similar to that in wild type (Fig. 7, J, N–P, R, and V–X). In contrast, reduced pav mutants exhibit significantly disorganized and diffuse actin mesh, consistent with Pav, but not Tum, enrichment at the oocyte cortex (Fig. 7, K–M and R–U).
As might be expected from their disrupted cortical actin, ooplasmic actin aggregates, and disrupted ooplasm actin mesh, reduced pav mutants exhibit premature ooplasmic streaming (Fig. 7, Z–CC’ and HH; and Video 6). Interestingly, tum RNAi oocytes do not exhibit premature ooplasmic streaming, consistent with retaining an ooplasm actin mesh similar to that in wild-type oocytes and displaying only subtle cortical actin alterations (Fig. 7, DD–FF’ and HH; and Video 6). Taken together with the ring canal findings, localization and mutant analyses indicate that Pav has Tum-independent, actin-related functions during normal oogenesis.
Reduced pav, but not tum, mutants exhibit premature ooplasmic streaming during oogenesis. (A–G) Time-lapse movies of stage 7 oocytes shown in Fig. 7: control (A), wimp/+ (B), reduced pavB200 (C), reduced pav963 (D), tumRNAi(1) (E), tumRNAi(2) (F), tumRNAi(2)+wimp (G), and PavDEAD (H). Time is indicated. Playback rate is 25 frames/s.
To further delineate Pav’s actin-dependent functions, we examined the effect of expressing the PavDEAD point mutation (Minestrini et al., 2002, 2003) using a maternal GAL4 driver on actin dynamics in oogenesis and cell wound repair. In oogenesis, PavDEAD exhibits a disorganized outer actin ring, disrupted actin mesh, and premature ooplasmic streaming, similar to that observed in reduced pav mutants; however, PavDEAD cortical actin phenotypes are less severe than reduced pav mutants (Fig. 7, A, I, Q, Y, GG, and HH; Fig. S4, J and R; and Video 6). We also examined PavDEAD phenotypes in colchicine-injected embryos to delineate the actin-dependent function of Pav in cell wound repair. Because PavDEAD affects the organization of MTs and the disruption of MTs impairs wound repair, we expected that PavDEAD would impair cell wound repair. Colchicine-injected embryos and PavDEAD exhibit similar phenotypes, delayed actin accumulation, and a slower closure rate (Fig. 8, A–C’ and E–G; and Video 7). Interestingly, PavDEAD with colchicine injection exhibits more severe phenotypes compared with colchicine-injected embryos alone: actin accumulates inside of wounds, slight wound overexpansion occurs, and they exhibit a markedly slower closure rate (Fig. 8, C–G; and Video 7). Thus, our results indicate that the actin bundling function of Pav is required for normal wound repair and oogenesis (Fig. 9).

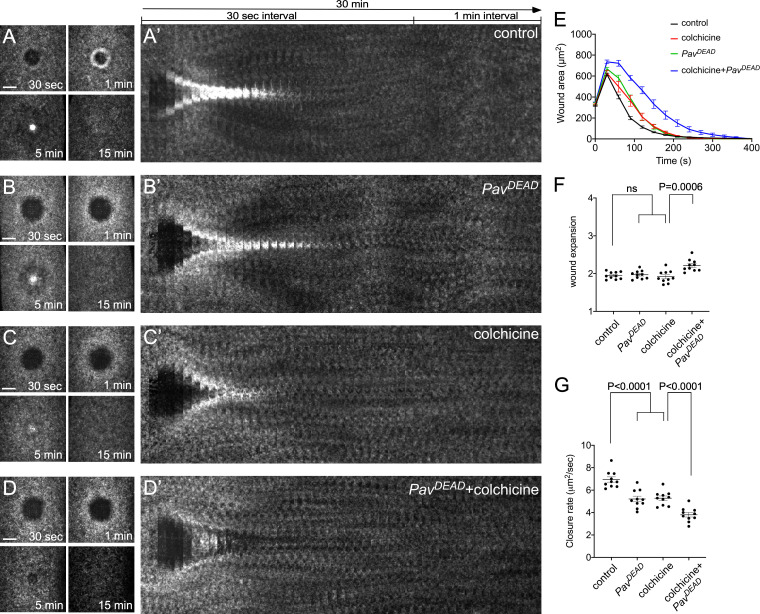
Actin-dependent Pav function is required for cell wound repair. (A–D) Actin dynamics (sGMCA or sStMCA) during cell wound repair in NC4–6 staged embryos: control (injection buffer alone; A), PavDEAD (B), colchicine injection (C), PavDEAD with colchicine injection (D). (A’–D’) xy kymograph across the wound area depicted in A–D, respectively. (E) Quantification of the wound area over time for control (injection buffer alone; n = 10), PavDEAD (n = 10), colchicine injection (n = 10), and PavDEAD with colchicine injection (n = 10). (F and G) Quantification of wound expansion time (F) and wound closure speed (G), control (buffer injection), PavDEAD, colchicine injection, and PavDEAD with colchicine injection (n = 10 for each condition). Time after wounding is indicated. Scale bar: 20 µm. Error bars represent ± SEM. Unpaired Student’s t tests were performed in F and G.
Actin bundling activity of Pav is required for cell wound repair. (A–G) Time-lapse confocal xy images from Drosophila NC4–6 staged embryos expressing an actin marker (sGMCA or sStMCA): control (buffer injection; A), PavDEAD (B), colchicine injection (C), PavDEAD + colchicine injection (D).

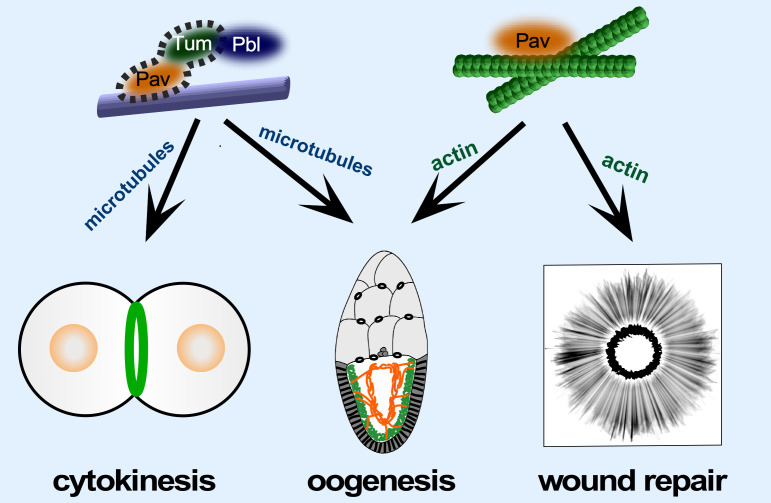
Model depicting the different roles of Pav and the centralspindlin complex in cytokinesis, oogenesis, and cell wound repair. Pav and Tum, as the centralspindlin complex, exhibit MT-dependent functions during cytokinesis. Pav, working independently of Tum, binds to actin during cell wound repair. Both Centralspindlin complex–dependent and –independent functions of Pav are required for normal oogenesis.
Pav is a kinesin-like protein that, along with the Tum RhoGAP protein, forms the centralspindlin complex, which both bundles and moves along MTs during cytokinesis (D’Avino et al., 2015; Green et al., 2012; Pollard and O’Shaughnessy, 2019; White and Glotzer, 2012). Mammalian CHO1/MKLP1 was previously shown to bind actin; however, the biological relevance of this activity was not reported (Kuriyama et al., 2002). Here, we show that Pav binds directly to F-actin through an entirely different region of the protein than CHO1/MKLP1, and that actin-dependent, centralspindlin-independent functions of Pav are required for proper cell wound repair and oogenesis. These findings have important implications for understanding the biological functions of Pav, as well as the processes of cell wound repair and oogenesis.
Cytoskeletal elements must be temporally and spatially coordinated for cells to carry out complex functions, including cell division and cell wound repair (Abreu-Blanco et al., 2011b; Agarwal and Zaidel-Bar, 2019; Basant and Glotzer, 2018; Bement and von Dassow, 2014; Chew et al., 2017; Chugh and Paluch, 2018; D’Avino et al., 2015; Dekraker et al., 2018; Dogterom and Koenderink, 2019; Green et al., 2012; Nakamura et al., 2018; Pollard and O’Shaughnessy, 2019; Sonnemann and Bement, 2011; Verboon and Parkhurst, 2015). Interestingly, we find that Pav localizes at the wound edge, overlapping with the innermost portion of the actin ring. Pharmacological disruption of this actin network disrupts Pav distribution. Previously, we found that Rho1 GTPase and the formin Diaphanous (Dia) similarly accumulate inside of the actin ring during cell wound repair, where they are both required for actomyosin ring assembly and, importantly, are not disrupted in the Pav mutant. One possible function for Pav in cell wound repair may be in organizing newly polymerized actin to generate a robust actomyosin ring and/or providing a mechanism for cross-talk between actin and MTs.
In addition to cell wound repair, we found that localization patterns and mutant phenotypes of Pav and Tum in oocytes suggest a subset of independent functions for each. As ring canals are the result of incomplete cytokinesis, we expected that Pav and Tum would function together as the centralspindlin complex at these structures. As such, we were unsurprised to find that both Pav and Tum were enriched along with actin at inner ring canals, and the inner ring canal defects we observed may be due to disrupted centralspindlin complex during cytokinesis. In contrast, only Pav was enriched with actin at outer ring canals. Pav may be required to organize and compact actin and/or MTs in the outer ring canals, since reduced-pav mutants exhibit detached and disorganized actin, similar to that observed in wash mutants (Wash also bundles and cross-links actin/MTs; Liu et al., 2009; Verboon et al., 2018).
Pav also works independently from Tum to organize the oocyte cortex, and its reduction leads to aberrant actin mesh organization and premature ooplasmic streaming. In contrast to this, tum mutants exhibit only minor actin defects in the oocyte and do not exhibit premature ooplasmic streaming. Similar to Pav, Rho1 GTPase, as well as the branched actin nucleation factor Wash and the linear actin nucleation factors Capu (formin) and Spire, are enriched at the oocyte cortex, have been shown to regulate actin and MT organization/cross-talk at the oocyte cortex, and, when mutant, result in premature ooplasmic streaming in stage 7–8 oocytes (Liu et al., 2009; Rosales-Nieves et al., 2006; Verboon et al., 2018). It is striking, but unclear, why so many different actin regulatory proteins are required nonredundantly to regulate actin and/or MT dynamics at the oocyte cortex, thereby preventing premature ooplasmic streaming. One possibility is that Pav has a major function in stabilizing the F-actin network by bundling actin or cross-linking F-actin/MTs that have already been polymerized by Wash, Capu, or Spire, since a primary function of these proteins is to nucleate F-actin filaments. Consistent with this, we observed detached actin aggregates within the ooplasm in reduced pav mutants. Another possibility is that Pav might regulate Rho1 activity in this context, thereby disrupting downstream effectors. Premature ooplasmic streaming has been proposed to occur when an actin mesh present in stage 7 oocytes is disrupted such that MTs are free to stream (Dahlgaard et al., 2007; Lu et al., 2016; Wang and Riechmann, 2008). Consistent with this model, the actin mesh in reduced pav mutant oocytes is disrupted, whereas the actin mesh in tum mutant oocytes is indistinguishable from that of wild type.
Dimerization/oligimerization via the stalk domain in Pav and other kinesin proteins is essential for bundling MTs and moving on MTs (Davies et al., 2015; Endow et al., 2010; Hirokawa et al., 2009; Sommi et al., 2010). Indeed, we show that the head domain alone cannot bundle MTs, whereas the head + stalk domains are sufficient to bundle/cross-link actin and MTs. However, the Pav head domain alone can bundle actin, suggesting that actin and MT binding sites are independently located within the head domain, and at least two actin binding sites exist. While kinesin protein moves on MTs, the binding states of the head domain depend on the nucleotides (Endow et al., 2010; Hirokawa et al., 2009). A nucleotide-free state induced with a mutation on the ATP binding site exhibits rigor-like binding to MTs, and motor function is disrupted. Previous studies found that PavDEAD (G131E mutation) exhibits a rigor-like state, and overexpression of PavDEAD stabilizes MTs in vivo (Minestrini et al., 2002, 2003). We expected that PavDEAD would enhance the bundling activity of Pav; however, we found that it significantly decreases Pav’s bundling activity for actin and MTs. Because PavDEAD protein can still bind to actin and MTs, PavDEAD protein might prevent modifications and/or disassembly by coating MTs. Although we have described PavDEAD phenotypes during oogenesis and cell wound repair, further molecular/structural studies are needed to fully delineate the actin binding activity of Pav and its interplay with MT dynamics.
In summary, we find that Pav and Tum localization is only partially overlapping during cell wound repair and oogenesis, leading to the identification of centralspindlin-dependent and -independent functions required for these processes. Importantly, we also show an unexpected actin-dependent function for the kinesin-like protein Pav during both cell wound repair and oogenesis, suggesting that Pav can change modes between primarily actin- or primarily MT-dependent functions in vivo. We have shown that Tum presence is not the factor that distinguishes these two modes of action. Identification of other interacting proteins is a future priority to understand the molecular basis for Pav choosing its mode of action and its centralspindlin-dependent versus -independent functions, as well as how widespread its noncanonical functions are.
Reagents used in this study are described in Table S1.
Flies were cultured and crossed at 25°C on yeast–cornmeal–molasses–malt medium. Flies used in this study are described in Table S1. All fly stocks were treated with tetracycline and then tested by PCR to ensure that they did not harbor Wolbachia.
To knock down genes, RNAi lines were driven maternally using the GAL4-UAS system with P{matα4-GAL-VP16}V37 for driving Tum RNAi (Bloomington; 7063; Nakamura et al., 2017; and 67923) or P{w[+mC]=GAL4::VP16-nos.UTR}MVD1 for driving Pav RNAi and PavDEAD. Reduced pav embryos were obtained from trans-heterozygous females by crossing pav963 or pavB200 females to RpL140wimp males (Liu et al., 2009; Parkhurst and Ish-Horowicz, 1991; Verboon et al., 2018). Localization patterns and mutant analyses were performed at least twice from independent genetic crosses, and ≥10 embryos were examined unless otherwise noted. Images representing the average phenotype were selected for figures.
sStMCA was generated by replacing the mCherry in sChMCA with Scarlet-i (Bindels et al., 2017) using standard PCR and cloning techniques. Transgenics were generated as previously described (Nakamura et al., 2017).
NC4–6 Drosophila embryos were collected at 0–30 min at room temperature (22°C). Embryos were hand dechorionated, placed onto no. 1.5 coverslips coated with glue, and covered with Series 700 halocarbon oil (Halocarbon Products Corp.).
All wounds were generated with a pulsed nitrogen N2 Micropoint laser (Andor Technology) tuned to 435 nm and focused on the cortical surface of the embryo. A region of interest was selected in the lateral midsection of the embryo, and ablation was controlled by MetaMorph. On average, ablation time was <3 s, and time-lapse imaging was initiated immediately. Occasionally, a faint grid pattern of fluorescent dots is visible at the center of wounds that arises from damage to the vitelline membrane that covers embryos.
Pharmacological inhibitors were injected from the dorsal side into the center of NC4–6 staged Drosophila embryos, and laser wounding was performed 5 min after injection. The following inhibitors were used: LatB (0.5 mM; EMD) and colchicine (25 mM; Sigma-Aldrich). Colchicine was prepared in injection buffer (5 mM KCl and 0.1 mM NaP, pH 6.8). LatB was prepared in injection buffer plus 10% DMSO. Injection buffer plus 10% DMSO or injection buffer alone were used as control.
All imaging was performed at room temperature (22°C). For live imaging, the microscope used was a Revolution WD systems (Andor Technology) mounted on a Leica DMi8 (Leica Microsystems) with a 63×/1.4-NA objective lens and controlled by MetaMorph software. Images and videos were acquired with 488 and 561 nm, using an iXon Ultra 897 or 888 electron-multiplying charge coupled device camera (Andor Technology). All images for cell wound repair were 17–20-µm stacks/0.25-µm steps. For single color, images were acquired every 30 s for 15 min and then every 60 s for 25 min. For dual green and red colors, images were acquired every 45 s for 30–40 min. Live imaging for premature ooplasm streaming was performed as previously described (Verboon et al., 2018).
For bundling/cross-linking assays and fixed tissues, the microscope used was a Zeiss LSM 780 spectral confocal microscope (Carl Zeiss Microscopy) fitted with Zeiss 20×/0.8, 40×/1.4, and 63×/1.4 oil Plan-Apochromat objectives. FITC (Alexa Fluor 488) fluorescence was excited with the 488-nm line of an argon laser, and detection was between 498 and 560 nm. Red (Alexa Fluor 568) fluorescence was excited with the 561-nm line of a DPSS laser, and detection was 570–670 nm. Far-red (Phalloidin 633) fluorescence was excited with the 633-nm line of an argon laser, and detection was 570–670 nm. Pinhole was typically set to 1.0 Airy units. Confocal sections were acquired at 0.25–1.0-μm spacing. Superresolution images were acquired using an Airyscan detector in Super Resolution mode, and captured confocal images were then processed using the Airyscan Processing feature of Zen software provided by the manufacturer (Carl Zeiss Microscopy).
All images were analyzed with Fiji (Schindelin et al., 2012). Measurements of wound area were done manually. To generate xy kymographs, all time-lapse xy images were cropped to 5.8 µm × 94.9 µm, and each cropped image was lined up. To generate fluorescent profile plots in R, 10-pixel sections across the wound from a single embryo were generated using Fiji as described previously (Nakamura et al., 2017). The lines represent the averaged fluorescent intensity, and the gray area is the 95% confidence interval. Line profiles from the left to right correspond to the top to bottom of the images, unless otherwise noted.
Quantification of the width and average intensity of the actin ring, wound expansion, and closure rate was performed as follows. The width of the actin ring was calculated with two measurements, the ferret diameters of the outer and inner edges of the actin ring 90 s after wounding. Using these measurements, the width of the actin ring was calculated as (outer ferret diameter − inner ferret dimeter) / 2. The average intensity of the actin ring was calculated with two measurements. Instead of measuring ferret diameters, we measured area and integrated intensity in same the regions as described in ring width. Using these measurements, the average intensity in the actin ring was calculated as (outer integrated intensity − inner integrated intensity) / (outer area − inner area). To calculate relative intensity for unwounded (UW) time point, average intensity at UW was measured with 50 × 50 pixels at the center of embryos, and the average intensity of the actin ring at each time point was divided by the average intensity of UW. Wound expansion was calculated as maximum wound area/initial wound size. Closure rate was calculated with two time points: (1) tmax, the time to reach maximum wound area, and (2) t1/2, the time to reach 50–35% size of maximum wound, because the slope of the wound area curve changes after t1/2. Using these time points, average speed was calculated with (wound area at tmax − wound area at t1/2) / tmax − t1/2. For analysis of actin mesh organization, GLCM texture analysis with four parameters, variance/contrast (the intensity between a pixel and its neighbor), correlation (linear dependency), uniformity/energy (proficient order in a whole image), and homogeneity (the tightness of distribution; Haralick et al., 1973), was performed with Matlab (MathWorks). To quantify protein recruitment to wounds, we subtracted the fluorescent intensity of the prewounding time point from the 180-s postwounding image. We then measured the averaged fluorescent intensity from 10 pixel sections across the wound in the subtracted image using Fiji. Line profiles were plotted, and area under the curve was measured using Prism 7.0a (GraphPad; Nakamura et al., 2017). Generation of all graphs, Student’s t test, and Fisher’s exact test were performed with Prism 7.0a or Matlab.
GFP-Pav, Pav, GFP-PavDEAD, and PavDEAD cDNAs were amplified as 5′SalI-3′NotI fragments from Ubi-GFP-Pav or UASp-GFP-PavDEAD flies (Minestrini et al., 2002) and then cloned into a double-tag pGEX vector (GST and His; Liu et al., 2009). Pav-head (1–500 aa), PavDEAD-head (1–500 aa with G131E mutation), Pav-head + stalk (1–712 aa), Pav-stalk + tail (491–887 aa), and Pav-tail (691–887 aa) domains were amplified from a double-tag Pav vector. Tum cDNA was amplified as a 5′BamHI-3′NotI from a spaghetti squash–sfGFP-Tum construct and then cloned into a pET21 vector. Primers used for cloning are described in Table S1. Protein expression assays were performed as previously described (Rosales-Nieves et al., 2006). CapuFH2 protein purification was performed as previously described (Rosales-Nieves et al., 2006). For GFP-Pav, Pav, GFP-PavDEAD, PavDEAD, and Tum proteins, cells were lysed by sonication in lysis buffer (10 mM Hepes, pH 7.5, 1 M NaCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM ATP, and 1 mM DTT) with 1% Triton X-100, 50 mM imidazole, and complete protease inhibitor tablets (Roche). Lysates were centrifuged at 10,000 g for 30 min, and the supernatants were coupled to Fastflow nickel-sepharose (GE) for 3 h at 4°C. The matrix was washed three times with lysis buffer with 50 mM imidazole and eluted by lysis buffer with 1 M imidazole. All His elutions, except for Tum protein, were coupled to glutathione-sepharose 4B (GE) for 3 h at 4°C, washed with lysis buffer, and then eluted with lysis buffer with 20 mM reduced glutathione. To reconstitute Pav and Tum complex, each supernatant of centrifuged Pav and Tum lysates was mixed and incubated on ice for 3 h. After His purification described above, an elution was coupled to glutathione-sepharose 4B (GE) for 3 h at 4°C, washed with lysis buffer to remove unbound Tum protein, and then eluted with lysis buffer with 20 mM reduced glutathione. For Pav-head, PavDEAD-head, Pav-head + stalk, Pav-stalk + tail, and Pav-tail, lysis buffer 2 (10 mM Hepes, pH 7.5, 250 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM ATP, and 1 mM DTT) was used. PreScission protease (GE) was used to elute proteins instead of reduced glutathione. All proteins were dialyzed into lysis buffer 2 and then flash frozen.
Rabbit muscle actin (Cytoskeleton) was polymerized in polymerization buffer (10 mM Tris, pH 7.5, 50 mM KCl, 2 mM MgCl2, and 1 mM ATP) and then stabilized with Alexa Fluor 488– or 633–conjugated Phalloidin. MT was polymerized by mixing unlabeled bovine brain tubulin (Cytoskeleton) and rhodamine-tubulin (Cytoskeleton) in a ratio of 1:5 and then stabilizing with paclitaxel (Cytoskeleton). MTs and test proteins were incubated in binding buffer A (for CapuFH2; 80 mM Pipes, pH 7.0, 1 mM MgCl2, 1 mM EGTA, 2 µM paclitaxel, 4 U/100 µl Alexa Fluor 488– or 633–and conjugated Phalloidin) or binding buffer A (for other proteins; 10 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1 mM ATP, 2 µM paclitaxel, and 4 U/100 µl Alexa Fluor 488– or 633–conjugated Phalloidin) for 15 min at room temperature. F-actin was then added and incubated for 10 min. The mixture of protein, actin, and MTs was pipetted onto slides and then visualized by fluorescence microscopy.
We used a commercial kit (Actin Binding Protein Spin-Down Assay Biochem Kit: rabbit skeletal muscle actin, Cytoskeleton) and followed manufacturer’s instructions using an Airfuge (Beckman Coulter).
To generate ovary lysates, ovaries were homogenized in Hepes lysis buffer (20 mM Hepes, pH 7, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1% Triton X-100, 5% glycerol, and 1 mM DTT) with added protease and phosphatase inhibitors (Roche). Lysate was sonicated five times using a Sonic Dismembrator (Model 60; Thermo Fisher Scientific) at setting 5 with 10 s/pulse. The extract was centrifuged at 13,500 g for 10 min at 4°C, and then the supernatant was recovered. Western blotting was performed according to standard procedures. The following antibodies were used: anti-α-tubulin (12G10; 1:5,000; DSHB), anti-Pav (1:1,000; provided by Dr. David Glover, University of Cambridge, Cambridge, UK; Adams et al., 1998), and anti-Tum (2B6+1H5; 1:10; DSHB).
Total RNA was obtained from 100 embryos (0–30 min old) using TRIzol (Invitrogen). 1 µg of total RNA was used for reverse transcription with the iScript gDNA Clear cDNA Synthesis Kit (Bio-Rad). RT-PCR analysis was performed using the iTaq Universal SYBR Green Supermix (Bio-Rad) with two individual parent sets and two technical replicates on the CFX96TM Real Time PCR Detection System (Bio-Rad). GAPDH1 was used as a reference gene. Percentage knockdown was calculated using the ΔΔCq calculation method compared with control (GAL4 only). The same primer sets for tum and GAPDH1 as previously described (Nakamura et al., 2017) were used.
Female flies were fattened on yeast for 2 d, and then ovaries were dissected and fixed. After three washes with PBS plus 0.1% Triton X-100, ovaries were permeabilized in PBS plus 1% Triton X-100 at room temperature for 2 h. Ovaries were washed three times with PAT (1× PBS, 0.1% Tween-20, 1% BSA, and 0.05% azide) and then blocked in PAT at 4°C for 2 h. Antibodies were used at the following concentrations: mouse anti-GFP monoclonal (1:100; Roche), rabbit anti-Pav polyclonal (1:250; provided by Dr. David Glover; Adams et al., 1998). The ovaries were incubated for 4 h at 4°C, washed three times with XNS (1× PBS, 0.1% Tween-20, 0.1% BSA, and 4% normal goat serum) for 40 min each, and then incubated with Alexa Fluor 488– and 568–conjugated secondary antibodies (1:1,000; Invitrogen) overnight at 4°C. Ovaries were washed with PTW (1× PBS and 0.1% Tween-20), incubated with Alexa Fluor 633–conjugated Phalloidin at 0.005 U/µl (Molecular Probes/Invitrogen) at room temperature for 1 h, and then washed with PTW. Ovaries were dissected into individual ovarioles and then mounted on slides in Slowfade Gold (Invitrogen). A minimum of two biological replicates were performed for each condition.
After 1–2 min, wounded embryos are transferred into formaldehyde-saturated heptane and incubated for 40 min. The vitelline membrane was removed by hand, and the embryos were washed three times with PAT and blocked in PAT for 2 h at 4°C. Embryos were incubated with primary antibodies for 24 h at 4°C and washed three times with XNS (1× PBS, 0.1% Tween-20, 0.1% BSA, and 4% normal goat serum) for 40 min each. Embryos were then incubated with secondary antibodies overnight at 4°C. Embryos were washed with PTW, incubated with Alexa Fluor 568–conjugated Phalloidin at 0.005 U/µl (Molecular Probes/Invitrogen) at room temperature for 1 h, washed with PTW, and imaged.
Female flies were fattened on yeast for 2 d, and then ovaries were dissected into cold PBS. Ovaries were fixed using 1:6 fix/heptane for 10 min (fix = 16.7 mM KPO4, pH 6.8, 75 mM KCl, 25 mM NaCl, 3.3 mM MgCl2, and 6% formaldehyde). Ovaries were washed three times with PBS plus 0.1% Triton X-100, and then incubated in PBS plus 0.5% Triton X-100 and Alexa Fluor 568–conjugated Phalloidin at 0.005 U/µl at room temperature for 1 h. Ovaries were washed with PTW 10 times for 10 min each, dissected into individual ovarioles, and mounted on slides in Slowfade Gold with DAPI medium (Invitrogen). A minimum of two biological replicates were performed for each condition.
All statistical analysis was done using Prism 8 (GraphPad). Gene knockdowns were compared with the appropriate control, and statistical significance was calculated using Student’s t test (Fig. 2, L–O, Q, and S; Fig. 7, R–Y; Fig. 8, F and G; and Fig. S1 G) or Fisher’s exact test (Fig. 7, A and HH) with P < 0.01 considered significant.
Table S1 describes all fly lines and reagents used in this study. Fig. S1 shows Rho family GTPase recruitment to wounds in reduced pav mutant backgrounds. Fig. S2 shows localization of sfGFP-Tum following injection of buffer, colchicine, LatB, and colchicine + LatB and proteins used in this study. Fig. S3 shows the results from bundling assays with Tum, Pav, and CapuFH2. Fig. S4 shows Pav localization in the ring canals and ring canal phenotypes in controls, reduced pav mutants, and tum knockdowns. Video 1 shows protein dynamics of Pav and Tum during cell wound repair. Video 2 shows the protein localization of Rho family GTPases in control and reduced pav mutants during cell wound repair. Video 3 shows the actin and Pav/Tum dynamics in controls, reduced pav mutants, and tum knockdowns during cell wound repair. Video 4 shows the protein localization of MT and Tum upon injecting buffer, colchicine, LatB, and colchicine + LatB during cell wound repair. Video 5 shows the protein localization of Pav upon injecting buffer, colchicine, LatB, and colchicine + LatB during cell wound repair. Video 6 shows the premature ooplasmic streaming in controls, reduced pav mutants, and tum knockdowns during oogenesis. Video 7 shows the actin dynamics in control, PavDEAD, colchicine, and PavDEAD + colchicine during cell wound repair. Table S1 describes all fly lines and reagents used in this study.
We thank David Glover, Julien Dubrulle (Fred Hutchinson Cancer Research Center, Seattle, WA), Marcos Gonzalez-Gaitan (University of Geneva, Geneva, Switzerland), Vladimir Gelfand (Northwestern University, Chicago, IL), Andrew Dominguez (Fred Hutchinson Cancer Research Center), Parkhurst laboratory members, the Bloomington Stock Center, the Harvard Transgenic RNAi Project, and the Developmental Studies Hybridoma Bank for advice, antibodies, DNAs, flies, graphics, and other reagents used in this study.
This work was supported by National Institutes of Health grant GM111635 to S.M. Parkhurst and in part by National Cancer Institute Cancer Center Support Grant P30 CA015704 (Shared Resources). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The authors declare no competing financial interests.
Author contributions: M. Nakamura, J.M. Verboon, C.L. Prentiss, and S.M. Parkhurst performed experiments. M. Nakamura, J.M. Verboon, and S.M. Parkhurst contributed to the design and interpretation of the experiments and to the writing of the manuscript.