
Fabricating artificial spider silk fibers in bulk scale has been a major goal in materials science for centuries. Two main routes have emerged for making such fibers. One method uses biomimetics in which the spider silk proteins (spidroins) are produced under nativelike conditions and then spun into fibers in a process that captures the natural, complex molecular mechanisms. However, these fibers do not yet match the mechanical properties of native silk fibers, potentially due to the small size of the designed spidroin used. The second route builds on biotechnological progress that enables production of large spidroins that can be spun into fibers by using organic solvents. With this approach, fibers that equal the native material in terms of mechanical properties can be manufactured, but the yields are too low for economically sustainable production. Hence, the need for new ideas is urgent. Herein, we introduce a structural-biology-based approach for engineering artificial spidroins that circumvents the laws with which spidroins, being secretory proteins, have to comply in order to avoid membrane insertion and provide a road map to the production of biomimetic silk fibers with improved mechanical properties.
Spiders have developed during the last 400 million years of evolution the skills to make the toughest fiber found in nature.1,2 They produce their fiber in fractions of a second, under ambient conditions, and from renewable resources, and still, its mechanical properties outperforms man-made materials like Kevlar.2 The unique and impressive mechanical properties of the fiber make it attractive for many different applications, e.g., for making high-performance textiles and sport goods,3 durable components for robotics,4 ropes and reinforcements of composite materials, and even for applications in medicine.3 Spider silk enhances wound healing5,6 and has successfully been used to bridge critical size nerve defects7,8 and as fascia replacements in animal models,9 further attesting to its potential usefulness.
Farming spiders and reeling silk is tedious, and for economically sustainable production, the spider silk proteins (spidroins) must be produced in heterologous hosts. For this purpose, a variety of hosts have been employed, e.g., prokaryotes, eukaryotic cell expression systems, plants, and even transgenic animals.10 However, the yields are typically poor, and the water solubility of the produced spidroins is low,11 which is probably why the vast majority of current protocols for production and spinning of recombinant spidroins include harsh solvents and denaturing agents.11 Fibers spun in this manner will be composed of aggregated proteins and thus likely have a hierarchal structure that differs from that of the native fiber, which is formed by spidroins that assemble into specific nanosized structures in a fine-tuned molecular process (cf. below).12 Expression and purification of spidroins under benign conditions, as well as subsequent biomimetic spinning protocols, have been developed and shown to recapitulate important events of the assembly process.13−16 However, the mechanical properties of fibers produced in this manner are still inferior to those of the native silk fiber, likely due to the small size of the spidroins used; hence, there is a need for novel ideas on how to produce high-performance spider silk replicas using biotechnological tools.
An individual spider can produce up to seven types of silks, each with characteristic mechanical properties.2,17 These fibers are mainly composed of spidroins that share an overall architecture of globular terminal domains of ca. 110–130 amino acid residues bracketing a repetitive region that is approximately 10 times longer (Figure 1).18−21 The terminal domains are unique to spidroins and regulate silk solubility and fiber formation, while the repetitive regions confer the mechanical properties to the fibers.22 The strongest silk, the dragline, is composed primarily of major ampullate spidroins (MaSps). The typical MaSp repeat region is composed of iterated poly-Ala blocks interspersed by Gly-rich repeats (Figure 1).

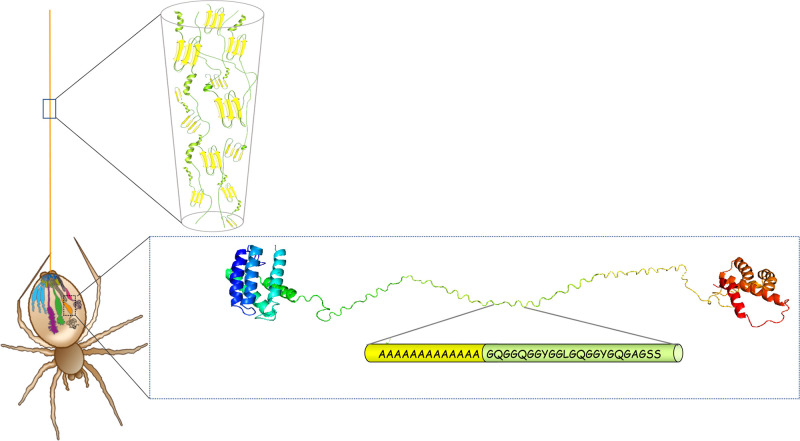
Schematic image of a spider’s silk glands and spidroin conformational states during storage and in the fiber. Spiders spin up to seven different kinds of silks from abdominal glands. The major ampullate gland is shown in orange and produces the dragline silk which is used as a life line. The enlargement of the major ampullate gland shows a typical major ampullate spidroin in a soluble state with an α-helical N-terminal domain (blue/cyan), a repetitive region in random coil conformation composed of many iterated poly-Ala and Gly-rich repeats (yellow/green), and an α-helical C-terminal domain (orange/red). The enlargement of the fiber shows the dominant protein structures in the fiber in which the Ala-blocks form β-sheets and β-crystals (yellow), whereas the Gly-rich repeats are unordered or adopt helical or β-turn conformations (green). The spider drawing is reproduced with permission from ref (32). Copyright 2015 Springer Nature.
In the silk gland, the spidroins are kept at high concentration (around 50% w/v).23 It is not fully elucidated how the spider manages to keep the spidroins at this extreme concentration without premature aggregation. Structural studies using nuclear magnetic resonance (NMR) spectroscopy on gland contents have shown that the repetitive region adopts mainly a random coil conformation during storage.24−26 The terminal domains form α-helix bundles and contribute to the hydrophilicity of the spidroins; these domains may also make up the outer shell of micelles during storage, shielding the repetitive entangled region in their core.27 Fiber formation takes place in the duct, in which a decreasing pH and increasing shear forces derived from pultrusion of the fiber lead to structural transitions of the spidroins.28,29 The N-terminal domain is monomeric during storage in the silk gland, whereas the C-terminal domain is dimeric, meaning that the spidroins are stored as “symmetrical” dimers. In response to lowered pH in the duct, the N-terminal domains dimerize, which locks the spidroins in a network and possibly cross-links the micelles.30,31 At the same time, shearing and acidification cause the C-terminal domain to unfold and to form β-sheet nuclei,21,28 which may trigger the transition of the repeat region into β-sheet conformation, in analogy to the seeding phenomenon in amyloid fibril formation.28 Collectively, this “lock and trigger” mechanism leads to conversion of the spidroin solution into a solid fiber.32
The dragline silk fiber is heterogeneous at the nanoscale level, with crystalline domains composed of stacked β-sheets embedded in an amorphous matrix (Figure 1). Despite its name, the amorphous phase contains specific secondary structures in which Gly-Pro-Gly-X-X motifs form type II β-turns, and Gly-Gly-X motifs form ordered structures with conformational similarities to collagen, polyproline helices,33,34 and possibly also short 310 helices.35 The bulk of the β-sheets are formed by the poly-Ala blocks, and the β-strands align with the fiber axis.25,33,36−39 The Ala methyl side chains enable β-strands from neighboring sheets to zip together. Poly-Ala β-sheet crystals will, thus, have methyl groups protruding into the void near the α carbons of the neighboring sheet (see below), resulting in tight enough packing to prevent water from entering the crystals,40 in a similar manner to “dry” steric zippers of amyloid crystals that are so tightly interdigitated that water cannot enter.41
Stretching a dragline silk fiber until failure results in distinct structural changes: First, stretching the amorphous regions leads to a yielding point, after which hydrogen bonds in the helices and β-turns break, leading to softening of the material. Upon further stretching, the fiber enters a phase where it is stiffened as the originally amorphous regions are completely extended, and the load is transferred to the β-sheet crystals. Finally, stick–slip deformation of β-sheet crystals leads to fiber failure.42−44 Thus, the heterogeneous structure of the dragline fiber (Figure 1) is key to its unique mechanical properties, and the crystalline domains ultimately confer the fiber’s high tensile strength.43 In agreement with this line of reasoning, increased crystallinity and higher degrees of order have been linked to higher tensile strength and stiffness and can be achieved by enhanced reeling speed and/or be conferred by spidroins with longer poly-Ala repeats.45−47
However, the exact relationships between crystallinity and mechanical properties remain to be elucidated; for example, molecular dynamic simulations have suggested that restricting the size of the crystalline domains is important for reaching the high tensile strength, resilience, and toughness of a spider silk fiber.43 The crystals act as intermolecular cross-linking points in a nanofishnet structure, which may enhance the toughness of silk filaments up to 1000 times compared to linear amyloid-like structures that lack cross-links.48 The relatively small crystals found in dragline silks mediate the required intermolecular contacts and enable the amorphous parts to extend fully until the β-sheet crystals are being stretched and eventually fail.43 In simulation studies, researchers have found that the optimal height of the stacked β-sheets is approximately 2–4 nm (corresponding to about 4–7 layers for poly-Ala) to mediate high tensile strength.43 The size restriction occurs because individual β-strands in small crystals can undergo stick–slip due to repeated breaking and reformation of hydrogen bonds, whereas higher stacks are apparently weaker because they are subject to bending and nonuniform tensile deformation of hydrogen bonds.44
Spidroins are secretory proteins, and as such, they are obliged to pass through the endoplasmic reticulum (ER) membrane to enter the secretory pathway. Proteins intended for secretion carry a signal peptide that directs the ribosome-nascent chain complex to the ER cytoplasmic surface.49 The ribosome docks to a translocon in the ER membrane, and the nascent chain is translated through the translocon into the ER lumen. The secretory protein is then transported to the Golgi apparatus before being exocytosed via vesicles that fuse with the plasma membrane. Instead of being translated across the translocon, the nascent chain can be cotranslationally inserted into the ER lipid membrane when the translocon opens up laterally,50 which happens if the nascent chain contains segments that, according to the biological hydrophobicity scale, promote membrane insertion.51,52
Thus, spidroins pass into the secretory pathway of the glandular cells and have to avoid being directed into the ER membrane during translation. This requirement precludes the use of stretches rich in Ile or Val because they unequivocally will be anchored in the lipid bilayer (Figure 2).51 Intriguingly, poly-Ala is the most nonpolar segment that will pass through the translocon without being membrane inserted, suggesting that MaSp repeat segments are optimized for hydrophobicity to the extent allowed by the translocon mechanism. Another structural biology constraint is that the preference to adopt the β-strand conformation correlates with hydrophobicity; the most β-prone residues are hydrophobic and, thus, mediate ER membrane insertion (Figure 2).53 Therefore, the requirement to translocate spidroins through the ER membrane precludes the spider from harnessing the most β-prone residues (Figure 2, green area of the graph), that is, exactly the side chains that would be expected to mediate the strongest intersheet interactions in β-crystals.

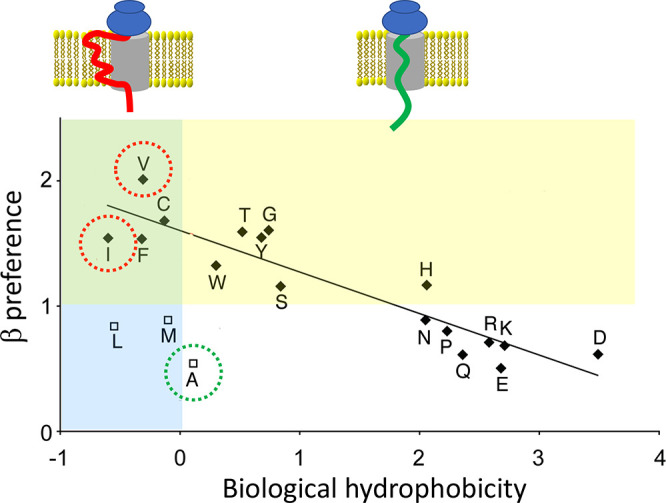
Membrane insertion is dictated by hydrophobicity. (upper part) Schematic image of the translocon (gray) with a transmembrane segment (red) and a secretory protein (green). (lower part) Correlation between biological hydrophobicity and preference to adopt the β-strand conformation over α-helix. The yellow area indicates amino acid residues that favor the β-strand conformation (values >1) whereas values <1 favor α-helix (see ref (53)). The blue area covers negative biological hydrophobicity values, which indicate residues that promote membrane insertion, whereas residues with positive values prevent membrane insertion (see ref (52)). The overlapping green area contains residues that both promote membrane insertion and favor β-strand conformation. Ala (green circle) prevents membrane insertion, and exchange of Ala for other aliphatic residues that favor the β-strand conformation (Val or Ile, red circles) will result in membrane insertion, which is incompatible with spiders’ spidroin production. The graph is reprinted with permission from ref (53). Copyright 2010 Elsevier.
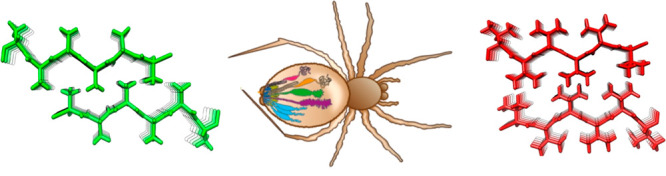
Close inspection of MaSp repetitive segments shows that they invariably contain poly-Ala segments. This finding suggests that poly-Ala, as opposed to segments composed of other residues, has evolutionary advantages and that the hydrophobic property of Ala (see Figure 2) is important for the formation of spider silk. This view raises an intriguing question as to whether repetitive segments composed of more hydrophobic residues (i.e., Val, Ile, Leu, Phe, Met, or Cys) would create even stronger silk but have been selected against because the use of these residues would trap the spidroins in the ER membrane (Figure 2). Generation of steric zipper predictions54 of poly-Val and poly-Ile shows that they will indeed form stable β-sheet interactions (Figure 3), whereas poly-Leu and poly-Phe will not. In support of this idea, amyloid fibrils in which hydrophobic side-chains form interstrand interactions are more stable than are fibrils with more polar buried residues.55,56 Poly-Cys or poly-Met are not attractive choices for making artificial silk because of their abilities to oxidize, and experiments show that introducing a few Cys in the poly-Ala segments does not give rise to substantially improved mechanical properties.57

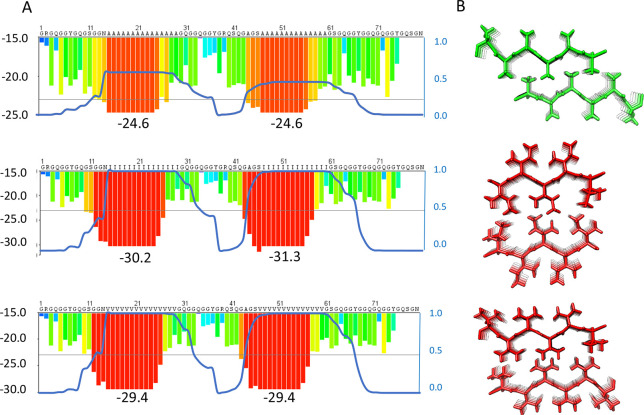
Optimization of spidroin repeat segments. (A) Examples of fibril energy profiles of a section of a repetitive region using the Zipper Database (ref (54)) for a wild type major ampullate spidroin (MaSp) repeat (top) and mutants in which the respective repeat sequence has been engineered to replace poly-Ala for poly-Ile (middle) or poly-Val (bottom). The Rosetta energies in kcal/mol of moving windows of hexapeptide steric zippers are indicated by the histograms with values on the left Y-axis and lowest numbers given below the histograms. Low Rosetta energies correlate to more stable steric zippers and, supposedly, stronger fibers. The blue lines show the probability of each residue to form a transmembrane segment using a scale between 0 and 1 as determined by the TMHMM v2.0 server at expasy.org (right Y-axis of each panel). Two transmembrane segments are predicted for the poly-Ile and poly-Val mutants, but the wild type is not predicted to insert as transmembrane helices although the poly-Ala blocks are on the border of predicted membrane insertion. (B) Steric zipper models of two β-sheets made up of Ala (top), Ile (middle), or Val (bottom).
The increased strength of β-sheet zippers composed of poly-Val or poly-Ile compared to poly-Ala, which, if extrapolated to spider silk β-crystals, would give stronger silks, reinforces the possibility that the former have been selected against for reasons other than silk properties. Predictions of the ability of MaSp repeat segments that have poly-Ala replaced with poly-Val or poly-Ile to form transmembrane helices confirm that they indeed will not pass through the translocon but will be inserted into the ER membrane (Figure 3). Other possible reasons for why spiders do not use poly-Ile or poly-Val are that these residues, in contrast to Ala, are essential,58 and that the lower aqueous solubility of poly-Val and poly-Ile compared to poly-Ala, even if they are short enough to pass through the translocon, may cause problems during the production and storage of spidroins.
Realizing that the nature of the eukaryotic secretory pathway may have hindered the evolution of optimal amino acid sequences for obtaining the strongest possible silk, we now propose to expand the sequence space of recombinant artificial spidroins produced in prokaryotic systems so that poly-Val, poly-Ile, and variants where such segments are combined with other sequences are investigated as well (Figure 4 and see below). Recombinant proteins expressed intracellularly in prokaryotes will avoid the requirement of passing through a lipid membrane, and importantly, Ile and Val are not essential amino acid residues for bacteria.58,59 Water solubility is a prerequisite for successful biomimetic spinning, and thus, the lower solubility of poly-Val and poly-Ile compared to poly-Ala may cause problems in future attempts to produce them in prokaryotic hosts. For the generation of artificial silk fibers using spinning methods based on organic solvents, the water solubility of the produced spidroins may be less important. However, if biomimetic spinning processes are desired, the required solubility could be achieved by one or several of the following strategies. First, recent results have shown that mini spidroins containing both terminal domains and a short repetitive region can efficiently be expressed in Escherichia coli and are extraordinarily soluble (50% w/v).13,15 Introducing Ala to Val or Ile substitutions in these proteins could therefore be afforded and still result in soluble target proteins. The mechanical properties of fibers spun from these proteins could match or even outperform those of native silk due to increased intermolecular interactions in the β-crystals. The dimensions of such mutant β-crystals would likely be similar to the native ones, which simulations have indicated are important for optimal mechanical properties.43,44 Second, if the replacement of all Ala residues results in insoluble products, variants with shorter poly-Val/Ile blocks as well as hybrid blocks containing two or more amino acid residue types could be designed. Third, the presence of a spidroin N-terminal domain may solve solubility problems for longer spidroins as well because the N-terminal domain and engineered variants thereof are exceptionally efficient in increasing solubility of aggregation-prone proteins of various sorts, including highly amyloidogenic polypeptides.20,60−64 Finally, engineering the Gly-rich repeats so that they compensate for the increased hydrophobicity of the β-sheet forming parts could be a means to improve overall solubility.

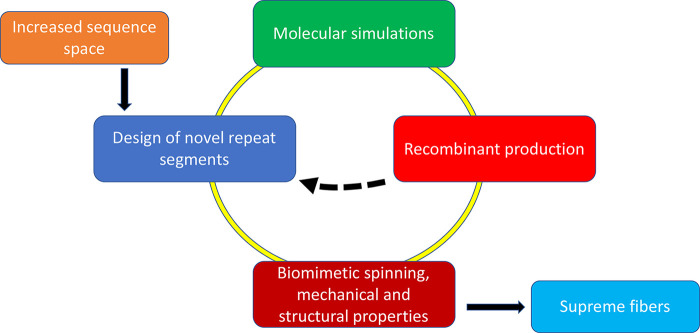
Road map to supreme artificial spider silk. Starting from the extended sequence space that becomes available by releasing the constraints imposed from spidroins having to pass through the eukaryotic secretory pathway, a multitude of potential repeat segments of artificial spidroins can be generated. Such segments can be screened for β-sheet stability by simulations and viable candidates passed for experimental screening of prokaryotic expression yields and water solubility, whereafter candidates with high production yields and water solubility can be spun into fibers. The dashed arrow indicates that candidates that show low yields or solubility can be fed back to inform on novel designs.
Recent sequencing of spider genomes shows that the spiders have a battery of silk genes (11 in Araneus ventricosus and 28 in Trichonephila clavipes), and the diversity in repeat regions between spidroins is remarkable.18,19 Harnessing this natural sequence space when designing variants of poly-Val and poly-Ile segments may further improve the chances of identifying artificial spidroin sequences that give optimal fiber properties. In addition, the composition of the silk fiber is more complex than previously thought, and silk production seems to depend on largely uncharacterized systems for chaperoning as well as ion homeostasis.18,65,66 Hence, stronger fibers may also be possible to generate by modulating the spinning methods as well as by tuning the protein composition, because composite dopes and fibers may outperform the single spidroin counterparts explored to date.
Based on these insights, we propose a strategy and road map for the production of novel artificial spidroins and superstrong silk fibers (Figure 4). In silico methods can be used to generate β-sheet crystals of poly-Val, poly-Ile, and variants thereof derived from the increased sequence space that becomes available from the insight that spiders cannot use long stretches of nonpolar residues for spidroin production. Such in silico β-sheet crystals can then be analyzed by molecular dynamics simulations for stability and strength, as has been done for naturally occurring segments.43,44,67 In addition to the novel repetitive segments that can be derived from the nonpolar segments that are not available to spiders, novel natural spidroin sequences are expected to become available from ongoing spider genome and transcriptome studies. The sequence space that can be generated by combining these two lines is vast, but still, molecular dynamics simulations will be able to provide valuable information on which designed repetitive segments are more likely to generate optimal fibers. However, simulations will not inform on properties of spidroins during recombinant production and purification why β-sheet design and simulations have to be revisited after experimental data have been generated on what artificial spidroins are possible to produce and spin into fibers. Iterated design–simulation–production–spin cycles (Figure 4) will enable the generation of high-performance and tailor-made biomimetic silk fibers optimized for specific purposes, by combining strength and other mechanical properties.
The approach outlined here will be important from a basic science point of view as well. The soluble form of designed spidroins can be evaluated by spectroscopic techniques and rheology in order to determine how they compare with natural feedstocks, and novel information on how different designed silk proteins fold, assemble, and mediate the fibers’ mechanical properties will increase our understanding of the natural spinning process as well as shed light on protein structure–activity relationships at large. Finally, we envision that testing not only poly-Val and poly-Ile (Figure 3) but also variants thereof that include novel naturally occurring sequences, which have different secondary structure propensities than the canonical poly-Ala in MaSps, in recombinant silk proteins can provide the information needed for deriving the exact relationships between the fibers’ structural and mechanical properties that are required to enable rational silk design in the future.
The authors declare no competing financial interest.
We are grateful to Michael Landreh and Gabriele Greco for valuable help with figures. This work was supported by European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement 815357), the Center for Innovative Medicine (CIMED) at Karolinska Institutet and Stockholm City Council, SFO Regen FOR 4-1364/2019, the Swedish Research Council (2019-01257 and 2016-01967), and Formas (2019-00427).