
- Altmetric
The mRNA modification N6‐methyladenosine (m6A) is associated with multiple roles in cell function and disease. The methyltransferases METTL3‐METTL14 and METTL16 act as “writers” for different target transcripts and sequence motifs. The modification is perceived by dedicated “reader” and “eraser” proteins, but not by polymerases. We report that METTL3‐14 shows remarkable cosubstrate promiscuity, enabling sequence‐specific internal labeling of RNA without additional guide RNAs. The transfer of ortho‐nitrobenzyl and 6‐nitropiperonyl groups allowed enzymatic photocaging of RNA in the consensus motif, which impaired polymerase‐catalyzed primer extension in a reversible manner. METTL16 was less promiscuous but suitable for chemo‐enzymatic labeling using different types of click chemistry. Since both enzymes act on distinct sequence motifs, their combination allowed orthogonal chemo‐enzymatic modification of different sites in a single RNA.
The N 6‐methyladenosine (m6A) methyltransferase METTL3‐METTL14 shows remarkable cosubstrate promiscuity, enabling sequence‐specific internal labeling of RNA. The transfer of ortho‐nitrobenzyl and 6‐nitropiperonyl groups allows enzymatic photocaging of RNA. METTL16 is less promiscuous but suitable for chemo‐enzymatic labeling via click chemistry. Since both enzymes act on distinct sequence motifs, their combination enables the modification of different sites in a single RNA.
Introduction
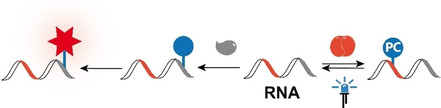
Covalent modifications of RNA represent the basis of numerous approaches aiming to make RNA accessible to biophysical and cellular studies by fluorescent, FRET, spin or biotin labeling. [1] Applications range from investigating the functional interplay of domains to studying transcriptional dynamics and tracking the localization in cells or in vivo. [2] The installation of photocleavable groups aims to control functions of RNA. [3] Ideally, labels should not interfere with the natural function and photocleavable groups should release the native biomolecule after irradiation, giving preference to modifications installed directly at the RNA without the need for additional tags.
Solid phase synthesis provides the most comprehensive toolbox for installation of functional groups in RNA and does not face limitations with respect to sequence or structural elements. [4] However, the size is limited to approximately 100 nt and long RNAs require chemical or enzymatic ligation of the synthetic fragment with a polymerase‐derived RNA. [5] Chemo‐enzymatic methods do not face length limitations and can be based on co‐ or post‐transcriptional introduction of modifications. While co‐transcriptional labeling was originally nucleoside‐ but not position‐specific, the development of unnatural base pairs has greatly expanded the possibilities but is still tedious to realize.[ 2f , 6 ] The use of specific polymerases like poly(A)‐polymerase can also provide position‐specificity. [7] Post‐transcriptional modifications have proven useful for labeling larger RNAs and even been applied in the cellular context. Notable examples are tRNA‐guanine transglycosylases (TGT), tRNAIle2‐agmatidine synthetase (Tias), and various approaches with methyltransferases (MTases). [8] However, all of these required a minimal structure motif that had to be appended to the RNA or acted on all RNAs with a specific feature, such as the 5′ cap or a free 2′‐OH group at the 3′ end.[ 8 , 9 ] Sequence‐specific modification required a guide‐RNA in addition to the MTase. [8k] Nucleic acid enzymes have been selected for site‐specific post‐transcriptional labeling of RNAs at the 2′ position or 3′ end by conjugating an additional appropriately labeled substrate, typically derived from an NTP analogue. [10] The recently described RNA acylation at induced loops (RAIL) approach allows to site‐selectively functionalize RNA via DNA‐induced structure, but faces limitations for functionalizing very short RNAs. [11]
For RNA photocaging, the same concepts and limitations apply. Here, the introduction of photocaging groups has been largely limited to chemical synthesis whereas post‐transcriptional enzymatic modification was dependent on tags.[ 3a , 12 ] We and others recently introduced MTases as tools to photocage DNA sequence‐specifically, [13] but enzymatic sequence‐specific photocaging of RNA sequences—be it by protein‐ or nucleic acid‐based enzymes—has not been reported to the best of our knowledge.
The recent rise of epitranscriptomics has led to reports on mRNA modifications and the responsible methyltransferases, including their recombinant production and crystal structures [14] (Figure S1). N 6‐methyladenosine (m6A) is the most prevalent internal modification in eukaryotic mRNAs and has been linked to fundamental biological processes, like neuronal development, [15] cell differentiation, [16] the circadian cycle [17] and diseases, including cancer. [18] In contrast to small nucleolar ribonucleoproteins (snoRNPs) and tRNA methyltransferases, the mRNA methyltransferases seem to require no additional guide RNA and nor rely on pre‐existing modifications. These enzymes might therefore be ideal for labeling mRNAs without the need to attach sequence or structure motifs and without the requirement of additional guide RNAs, but this potential has not been explored to date.
Results and Discussion
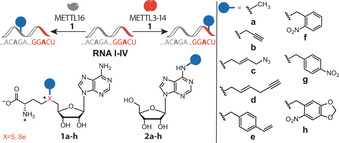
We chose to assess the potential of the two currently known mRNA methyltransferases responsible for m6A formation, METTL3‐METTL14 (abbreviated as METTL3‐14) and METTL16, for sequence‐specific labeling and photocaging of RNAs (Scheme 1). METTL3‐14 is a heterodimer with preference for the DRACH motif (Figure S1 A).[ 14 , 19 ] The available crystal structures of SAM‐bound METTL3‐14 in pre‐catalytic state suggest that the AdoMet analogs could fit into the free space in the binding pocket (Figure S1 A). In cells, METTL3‐14 functions in complex with other proteins, [20] but the heterodimer alone is active in vitro and able to transfer allyl and propargyl groups from analogs of the natural cosubstrate S‐adenosyl‐l‐methionine (AdoMet). [21] METTL16 requires a different consensus motif (UACAGAGAA) that needs to be presented as a loop in a hairpin structure. [22] METTL16 was shown to methylate only two target RNAs, but additional target transcripts are likely. [22] Neither of these enzymes requires an additional guide RNA to target the consensus motif.

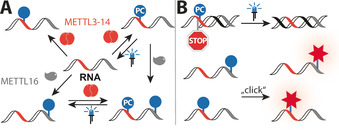
A) Methyltransferases METTL3‐14 and METTL16 used for sequence‐specific modification of RNA without dedicated tags. B) Photocaging (PC) groups block reverse transcription in a reversible manner. Bioorthogonal groups can be used for RNA labeling using different types of click chemistry.
To explore the cosubstrate scope of METTL3‐14 and METTL16, we designed three different substrate RNAs containing the respective consensus motifs (I–III, Table S1). METTL3‐14 was tested with RNA I, containing four repeats of the GGACU consensus motive. Using RNA I and AdoMet (1 a) resulted in 55 % of m6A, according to HPLC analysis after digestion to single nucleosides (Table 1). SeAdoYn (Se‐1 b) as cosubstrate yielded 45 % of 2 b, corresponding to 82 % relative to AdoMet. AdoMet analogs with longer alkyl chains like azidobutenyl‐ (1 c) or hexenynyl‐ (1 d) were not efficiently converted. However, the benzylic AdoMet analogs with vinylbenzyl‐ (1 e) and ortho‐nitrobenzyl‐ (1 f) groups (ONB) gave high relative conversions (58 % and 55 % compared to AdoMet). Moreover, we synthesized a new AdoMet analog carrying a 6‐nitropiperonyl (NP) group (1 h), which gave a 53 % conversion relative to AdoMet. All N 6‐modified adenosine products were confirmed by LC‐MS analysis (Figure S2). Controls using heat‐inactivated enzyme did not yield product peaks (Figure S3). The kinetics of the transfer reactions indicated that, as expected, natural AdoMet is a better cosubstrate than all AdoMet analogs tested (Figure S4 and Table S4). Nevertheless, the data summarized in Table 1 show that METTL3‐14 is able to transfer short alkyl and benzylic groups to the N 6‐position of adenosine making RNA with the consensus motif GGACU accessible to sequence‐specific labeling and photocaging.

|
Entry |
Substrate |
Theoretical Yield [%] |
RNA |
MTase |
Product |
A modification [%] |
Relative modification [%] |
Site‐ specific modification A [%] |
|---|---|---|---|---|---|---|---|---|
|
1 |
1 a |
100 |
I |
3‐14 |
2 a |
55 |
100 |
55 |
|
2 |
Se‐1 b |
100 |
I |
3‐14 |
2 b |
45 |
82 |
45 |
|
3 |
1 c |
100 |
I |
3‐14 |
2 c |
tr. |
tr. |
– |
|
4 |
1 d |
100 |
I |
3‐14 |
2 d |
tr. |
tr. |
– |
|
5 |
1 e |
100 |
I |
3‐14 |
2 e |
32 |
58 |
32 |
|
6 |
1 f |
100 |
I |
3‐14 |
2 f |
30 |
55 |
30 |
|
7 |
1 g |
100 |
I |
3‐14 |
2 g |
tr. |
tr. |
– |
|
8 |
1 h |
100 |
I |
3‐14 |
2 h |
29 |
53 |
29 |
|
9 |
1 a |
5.9 |
II |
16 |
2 a |
3.6 |
100 |
61 |
|
10 |
Se‐1 b |
5.9 |
II |
16 |
2 b |
2.5 |
69 |
42 |
|
11 |
1 c |
5.9 |
II |
16 |
2 c |
tr. |
tr. |
– |
|
12 |
1 f |
5.9 |
II |
16 |
2 f |
n.d. |
n.d. |
n.d. |
|
13 |
1 a |
14.3 |
III |
16 |
2 a |
7.0 |
100 |
49 |
|
14 |
Se‐1 b |
14.3 |
III |
16 |
2 b |
2.8 |
40 |
19 |
|
15 |
1 c |
14.3 |
III |
16 |
2 c |
tr. |
tr. |
– |
|
16 |
1 d |
14.3 |
III |
16 |
2 d |
tr. |
tr. |
– |
|
17 |
1 f |
14.3 |
III |
16 |
2 f |
n.d. |
n.d. |
– |
|
18 |
1 g |
14.3 |
III |
16 |
2 g |
n.d. |
n.d. |
– |
Next, we tested the cosubstrate scope of METTL16, recombinantly expressed in insect cells (Figure S5), using RNAs of different lengths containing the target sequence (RNA II: 61 nt and RNA III: 26 nt).[ 22b , 23 ] These RNAs contain 17 and 7 adenosines, respectively, but only a single target adenosine each. Therefore, the total amount of site‐specifically modified A would correspond to 14.3 % or 5.9 %, respectively (Table 1).
Using the natural cosubstrate (1 a) on RNA II or III yielded 61 or 49 % of site‐specifically methylated m6A, respectively (Table 1), with the longer RNA II being a better substrate than RNA III. Using SeAdoYn (Se‐1 b) yielded 42 % and 19 % of site‐specifically modified 2 b for RNA II and III, respectively (Figure S6), corresponding to 40–69 % relative yield compared to AdoMet. In addition, METTL16 was able to transfer the 4‐azido‐but‐2‐enyl group from 1 c yielding the corresponding product 2 c, albeit in low yields. All N 6‐modified products were confirmed by LC‐MS (Figure S6). Control reactions with inactivated enzyme did not yield products (Figure S7). The kinetics of the transfer reactions were measured (Figure S4 and Table S4). In contrast to METTL3‐14, METTL16 did not transfer benzylic groups (Table 1). These data show that METTL16 can transfer longer alkyl chains including bioorthogonal groups to the sequence motif UACAGAGAA, which is distinct from METTL3‐14. However, its promiscuity does not comprise benzylic groups. In contrast to METTL3‐14, there are no crystal structures available for SAM‐bound METTL16 in pre‐catalytic state, so its substrate promiscuity cannot be readily assessed from the available structural data (Figure S1 B).
To assess whether the post‐synthetically modified RNAs I–III could be further functionalized, we used the copper‐catalyzed (CuAAC) and strain‐promoted (SPAAC) azide‐alkyne cycloaddition for functionalization with Cy5 as a fluorescent dye or biotin as an affinity handle (Figure 1). Successful clicking of the N 6‐propargylated RNA I was confirmed by LC‐MS for biotin‐azide (Figure 1 B and Figure S8 A,B) and in‐gel fluorescence for Cy5‐azide, which also proved that the RNA remained intact during the click procedure (Figure 1 C, Figure S8 C). Similarly, RNA with N 6‐(azido‐but‐2‐enyl)‐adenosine was labeled via SPAAC using DBCO‐Cy5, whereas controls with heat inactivated enzymes were not (Figure S9). Since every long RNA is likely to contain DRACH motives, we used METTL3‐14 to label mRNA produced by in vitro transcription (IVT) coding for the reporter protein firefly luciferase (FLuc mRNA, ≈1900 nt long) with Cy5 azide (Figure S10).

![A) CuAAC of RNA I containing N
6‐propargyl‐adenosine from enzymatic modification via METTL3‐14 using Cy5‐azide or biotin‐azide. B) LC‐QTOF‐MS analysis of RNA after enzymatic digestion to ribonucleosides before and after CuAAC using biotin‐azide. Left: EIC of N
6‐propargyladenosine (2 b, C13H15N5O4 [M+H]+ 306.120±0.005), right: Extracted ion chromatogram (EIC) for N
6‐biotinylated adenosine (2 i, C26H37N11O6S [M+H]+ 632.272±0.005, also see Figure S8 A). C) PAA gel analysis of RNA after CuAAC with Cy5‐azide on SybrGold and Cy5 channels (lane 1). Control contained heat‐inactivated enzyme (lane 2).](/dataresources/secured/content-1765906199534-57b71c74-2d2d-443e-997b-2ad82c5fb8e2/assets/ANIE-60-4098-g001.jpg)
A) CuAAC of RNA I containing N 6‐propargyl‐adenosine from enzymatic modification via METTL3‐14 using Cy5‐azide or biotin‐azide. B) LC‐QTOF‐MS analysis of RNA after enzymatic digestion to ribonucleosides before and after CuAAC using biotin‐azide. Left: EIC of N 6‐propargyladenosine (2 b, C13H15N5O4 [M+H]+ 306.120±0.005), right: Extracted ion chromatogram (EIC) for N 6‐biotinylated adenosine (2 i, C26H37N11O6S [M+H]+ 632.272±0.005, also see Figure S8 A). C) PAA gel analysis of RNA after CuAAC with Cy5‐azide on SybrGold and Cy5 channels (lane 1). Control contained heat‐inactivated enzyme (lane 2).
When biotin‐azide was clicked to the enzymatically propargylated RNA, we observed strong termination in a reverse transcription assay (Figure S11). Based on our finding that METTL3‐14 accepts 1 f and 1 h and transfers the ONB and NP groups to the N 6‐position of adenosine, we sought to explore the photocaging/‐uncaging ability for the respective RNA (Figure 2 A). While MTase‐based transfer and removal of photocaging groups has been reported previously for DNA, [13a] enzymatic transfer and light‐triggered removal of the ONB or NP derivatives was not reported for RNA to date. Here, irradiation of RNAs containing the ONB group installed by METTL3‐14 completely removed the ONB group after 2 min at either 365 nm or 405 nm (Figure 2 B and Figure S12 A,B). The NP group was completely removed after 1 min irradiation at 420 nm, according to LC‐MS (Figure S12 C), in line with previous reports about the NP group for DNA photocaging. [24] Uncaging at higher wavelengths is desirable to reduce photo‐damage and phototoxicity, especially in a cellular context. Gel analysis showed that the RNA was not degraded by irradiation under used conditions (Figure 2 C and Figure S12 D,E). Importantly, after light‐triggered removal of the photocaging groups, the RNA could again be enzymatically remodified (Figure S13), further supporting intactness of RNA after irradiation.

![A) Scheme of photocleavage of the ONB group from enzymatically modified target RNA. B) Analysis of photocleavage shown in (A) using LC‐MS after digestion to ribonucleosides. EICs for 2 f are shown (C17H18N6O6 [M+H]+ 403.136±0.005). C) Gel analysis of samples from (A) before and after irradiation (15 % denaturing PAGE). D) Reverse transcription (RT) of RNA IV modified using METTL3‐14 and Se‐1 b or 1 f. For 1 f a new termination band is observed at 36 nt, 1 nt downstream of the modified A. This termination band is reduced if the RNA was irradiated before RT. Ctrl shows RT for unmodified RNA IV. Full gel is shown in Figure S14 B.](/dataresources/secured/content-1765906199534-57b71c74-2d2d-443e-997b-2ad82c5fb8e2/assets/ANIE-60-4098-g002.jpg)
A) Scheme of photocleavage of the ONB group from enzymatically modified target RNA. B) Analysis of photocleavage shown in (A) using LC‐MS after digestion to ribonucleosides. EICs for 2 f are shown (C17H18N6O6 [M+H]+ 403.136±0.005). C) Gel analysis of samples from (A) before and after irradiation (15 % denaturing PAGE). D) Reverse transcription (RT) of RNA IV modified using METTL3‐14 and Se‐1 b or 1 f. For 1 f a new termination band is observed at 36 nt, 1 nt downstream of the modified A. This termination band is reduced if the RNA was irradiated before RT. Ctrl shows RT for unmodified RNA IV. Full gel is shown in Figure S14 B.
Next, we were interested to see whether the ONB or NP groups at the N 6‐position of adenosine would affect a biological function. Recent work showed that N 6‐methylation of adenosine does not have significant effects on wild‐type polymerases, whereas extended residues such as triazoles or engineered polymerases impeded the reverse transcription.[ 21b , 25 ] We therefore designed a longer RNA containing two METTL3‐14 consensus motifs and a primer binding site for reverse transcription (Table S1). This RNA IV was made by IVT, modified post‐transcriptionally using Se‐1 b or 1 f, respectively, and then used for reverse transcription (RT) (Figure 2 D). In this experiment, RNA V served as a control, with GGACU motif replaced by GGCCU. A control RT with unmodified RNA revealed that out of the two GGACU consensus motifs, the one harbouring the target adenosine at 35 nt was suitable for analysis by RT because it did not show termination for unmodified control RNA (Figure S14). When RNA IV was treated with 1 f and METTL3‐14, the RT showed a termination band at 36 nt, corresponding to one nucleotide downstream of the modified adenosine. This band was almost absent when the sample was irradiated for 2 min at 365 nm or 405 nm prior to RT (Figure 2 D and Figures S14 and S16). According to the LC‐MS analysis, these irradiation conditions completely removed the ONB‐modifications and according to polyacrylamide (PAA) gel analysis they did not lead to degradation of the RNA (Figure 2 B,C and Figures S14–16). Similar results were observed when RNA IV was treated with 1 h and METTL3‐14 while the control RNA V showed no termination at 36 nt (Figure S16 A). In this case, irradiation for 1 min at 420 nm was sufficient to remove the NP modification, according to LC‐MS (Figure S16 B). Additionally, all modified RNAs contained a termination band at 34 nt, corresponding to one nucleotide upstream of the modification (Figure 2 D and Figures S14 and S16). These data show that the ONB and NP groups can be removed by brief irradiation with UV light without degrading the RNA. Placing the ONB/NP group at the N 6‐position of adenosine significantly increases termination opposite this modification. For the first time, a post‐synthetically installed ONB/NP group in RNA can be removed by light.
Since METTL3‐14 and METTL16 target distinct sequence motifs, it should be possible to sequence‐specifically label RNAs containing both target motifs. To assess this option, we designed a chimeric RNA containing both the METTL3‐14 and the METTL16 consensus sequences as well as control chimeras with point mutations in either the METTL3‐14 or the METTL16 target sites (Table S1).[ 14b , 26 ] As expected, these control chimeras were only modified by the MTase whose target motif remained intact, confirming specificity of the enzymes (Figure 3 A,B).

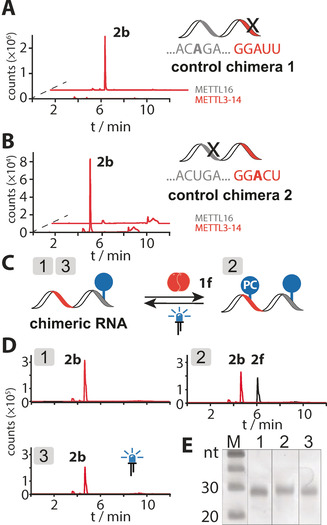
Orthogonal labeling approach of an RNA bearing METTL3‐14 and/or METTL16 consensus motifs (shown in red and gray, respectively). A) Enzymatic modification of RNA bearing a mutation in the METTL3‐14 consensus motif (control chimera 1). EICs for N 6‐propargyladenosine (2 b, m/z=306.1). B) Enzymatic modification of RNA bearing a mutation in the METTL16 consensus motive (control chimera 2). LC‐MS analysis as in (A). C) Scheme of the reversible double modification. D) LC‐QQQ‐MS analysis after each modification step. Extracted ion chromatograms for N 6‐propargyladenosine (2 b, m/z=306.1) and N 6‐(o‐nitrobenzyl)adenosine (2 f, m/z=403.1) shown in red and black, respectively. E) Analysis of the modified RNA on 15 % PAA gel.
Knowing that MTase‐based modification is sequence‐specific, we tested dual modification of the chimeric RNA by first applying Se‐1 b and METTL16 (step 1 in Figure 3 C/D) followed by treatment with 1 f and METTL3‐14 (step 2 in Figure 3 C/D). After each step, LC‐MS analysis confirmed that the modification was successful (Figure 3 D and Figure S17). Indeed, the double modification of the chimeric RNA was possible by sequential use of METTL16 and METTL3‐14 using different AdoMet analogs. Finally, we also showed that the transferred ONB group could be selectively removed by light from the double modified chimeric RNA (step 3 in Figure 3 C/D). During all steps, the RNA remained intact (Figure 3 E).
Conclusion
Taken together, our data show that mRNA methyltransfersases provide a means to label RNAs containing their motifs at internal sites. In the case of METTL3‐14, a ≈1900 nt FLuc‐mRNA could be labeled without the need to append a structural hallmark (like the 5′ cap) or secondary structure element (like for TGT or Tias). This is possible because the cosubstrate scope of human m6A MTases is broader than previously expected, especially for METTL3‐14, which even accepts photocaging groups. Looking at the crystal structures of METTL3‐14 in complex with AdoMet the accommodation of large substituents can be anticipated (Figure S1). The sequence‐specific chemo‐enzymatic labeling via propargylation and subsequent click chemistry is highly efficient. The transfer of photocaging groups is ≈50 % compared to AdoMet, which was sufficient to impair reverse transcription. Protein engineering should allow to further increase the yield and provide a way to control biological functions at exactly the sites, where modifications occur in nature. As a proof of concept, we showed that photocaging groups block primer extension during reverse transcription, but we anticipate effects on stability and binding of proteins and miRNAs, as previously shown for chemically photocaged short nucleic acids.[ 3a , 12a ] In particular, for studying the function of epitranscriptomic modifications, we envision that blocking MTase target sites and the interaction with their “reader” proteins could become a very attractive tool. Sequence‐specific labeling has been achieved previously with MTases but required the addition of a guide‐RNA complicating the system. [8k] The target requirements of METTL3‐14 and METTL16 are small, especially METTL3‐14 only requires a 5 nt consensus motif (DRACH) that leaves room for variations. Based on m6A sequencing, 12,000 METTL3‐14 target sites in over 7,000 genes were found in mammals, [27] suggesting that most mRNAs would be available for labeling/manipulation with this enzyme complex without the need to append a tag nor add a guide RNA. Finally, we provide proof of concept that the two MTases can be used for orthogonal labeling of RNA at different sites. This can be useful for double labeling (including FRET labels or fluorophore/quencher pairs for biophysical studies) or labeling an mRNA that should also become activated by light (e.g. to activate and track RNA).
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
A.R. gratefully acknowledges funding by the DFG (RE2796/6‐1, RE2796/7‐1) and the ERC (772280). A.O. is supported by the Graduate School of the Cells‐in‐Motion Cluster of Excellence (EXC 1003‐CiM), University of Münster, Germany. We thank S. Wulff, Dr. W. Dörner, Dr. P. Špaček, N. Muthmann, D. Reichert, K. Hartstock, A.‐M. Lawrence‐Dörner, F. Vinke and Dr. L. Anhäuser for excellent experimental assistance, Prof. Dr. Markus Bohnsack (University of Göttingen) for providing the METTL16‐encoding plasmid and Prof. Dr. Gunter Meister (University of Regensburg) for the pFastBacDual_METTL3_METTL14 plasmid in the framework of SPP1784. Open access funding enabled and organized by Projekt DEAL.
References
2
2a
2a
2c
2d
2e
2f
2f
2g
2h
3
3b
3b
5
5c
5c
6
6b
6c
7
8
8b
8c
8c
8d
8e
8f
8f
8g
8g
8h
8j
8j
8k
9
10
10a
10e
10e
12
12c
12c
12d
12e
12e
13
13a
13b
14
14a
16
16a
16b
17
17a
17b
18
18a
18b
18d
20
21
21a
21b
21b
22
22a
22b
22d
22e
23
24
25
25
26
26
27
27a
 Tag‐Free Internal RNA Labeling and Photocaging Based on mRNA Methyltransferases
Tag‐Free Internal RNA Labeling and Photocaging Based on mRNA Methyltransferases
